3ODG
 
 | | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis | | Descriptor: | CHLORIDE ION, XANTHINE, Xanthosine phosphorylase | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | crystal structure of xanthosine phosphorylase bound with xanthine from Yersinia pseudotuberculosis
To be Published
|
|
3OF3
 
 | | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type 1 | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of PNP with an inhibitor DADME_immH from Vibrio cholerae
To be Published
|
|
3OCC
 
 | | Crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Kim, J, Ramagopal, U.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | crystal structure of PNP with DADMEimmH from Yersinia pseudotuberculosis
To be Published
|
|
3OHP
 
 | |
3QNM
 
 | | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function | | Descriptor: | CHLORIDE ION, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Matthew, M.W, Ramagopal, U.A, Toro, R, Dickey, M, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-08 | | Release date: | 2011-03-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Haloalkane Dehalogenase Family Member from Bacteroides thetaiotaomicron of Unknown Function
To be Published
|
|
3RP1
 
 | | Crystal structure of Human LAIR-1 in C2 space group | | Descriptor: | Leukocyte-associated immunoglobulin-like receptor 1 | | Authors: | Sampathkumar, P, Ramagopal, U.A, Yan, Q, Toro, R, Nathenson, S, Bonanno, J, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Human LAIR-1 in C2 space group
To be Published
|
|
3RBG
 
 | | Crystal structure analysis of Class-I MHC restricted T-cell associated molecule | | Descriptor: | Cytotoxic and regulatory T-cell molecule, PHOSPHATE ION | | Authors: | Rubinstein, R, Ramagopal, U.A, Toro, R, Nathenson, S.G, Fiser, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2011-03-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional classification of immune regulatory proteins.
Structure, 21, 2013
|
|
3RNQ
 
 | |
3RNK
 
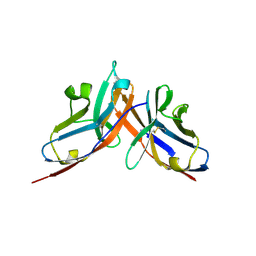 | | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain | | Descriptor: | Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Lazar-Molnar, E, Ramagopal, U.A, Cao, E, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the complex between mouse PD-1 mutant and PD-L2 IgV domain
To be Published
|
|
3GHY
 
 | | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2 | | Descriptor: | Ketopantoate reductase protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative ketopantoate reductase from Ralstonia solanacearum MolK2
To be Published
|
|
3G7U
 
 | | Crystal structure of putative DNA modification methyltransferase encoded within prophage Cp-933R (E.coli) | | Descriptor: | CHLORIDE ION, Cytosine-specific methyltransferase, GLYCEROL | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Gilmore, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-10 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of DNA Modification Methyltransferase Encoded within Prophage Cp-933R (E.coli)
To be Published
|
|
3GD5
 
 | | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Ramagopal, U.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Gloeobacter violaceus
To be Published
|
|
3GRZ
 
 | | CRYSTAL STRUCTURE OF ribosomal protein L11 methylase FROM Lactobacillus delbrueckii subsp. bulgaricus | | Descriptor: | GLYCEROL, Ribosomal protein L11 methyltransferase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-26 | | Release date: | 2009-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN 11 METHYLASE FROM Lactobacillus delbrueckii subsp. bulgaricus
To be Published
|
|
3GMF
 
 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3K12
 
 | | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, uncharacterized protein A6V7T0 | | Authors: | Ho, M, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of an uncharacterized protein A6V7T0 from Pseudomonas aeruginosa
To be published
|
|
2OB3
 
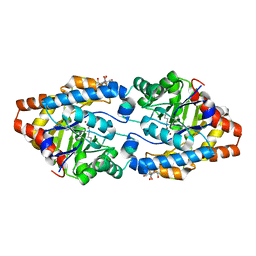 | | Structure of Phosphotriesterase mutant H257Y/L303T | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Parathion hydrolase, ZINC ION | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure of Phosphotriesterase mutant H257Y/L303T
To be Published
|
|
2O4M
 
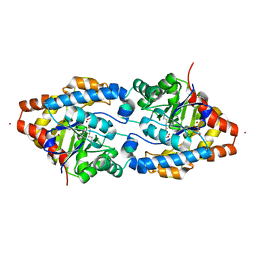 | | Structure of Phosphotriesterase mutant I106G/F132G/H257Y | | Descriptor: | ACETIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of Phosphotriesterase mutant I106G/F132G/H257Y
To be Published
|
|
2OYP
 
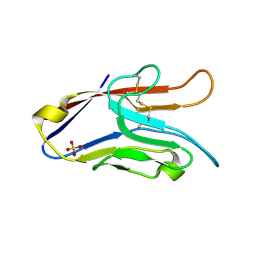 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
2Q1M
 
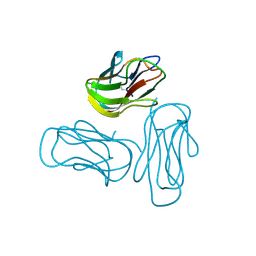 | |
2Q3N
 
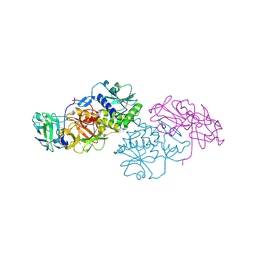 | | Agglutinin from Abrus Precatorius (APA-I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 A chain, Agglutinin-1 B chain | | Authors: | Bagaria, A, Surendranath, K, Ramagopal, U.A, Ramakumar, S, Karande, A.A. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Function Analysis and Insights into the Reduced Toxicity of Abrus precatorius Agglutinin I in Relation to Abrin.
J.Biol.Chem., 281, 2006
|
|
2Q91
 
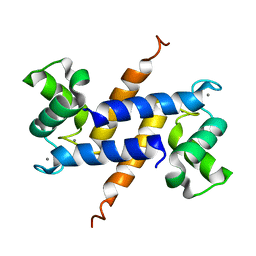 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
2QDN
 
 | |
2R32
 
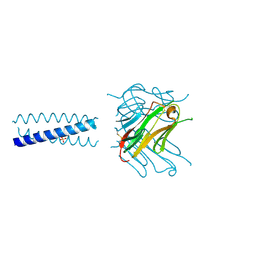 | | Crystal Structure of human GITRL variant | | Descriptor: | GCN4-pII/Tumor necrosis factor ligand superfamily member 18 fusion protein, SULFATE ION | | Authors: | Chattopadhyay, K, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Assembly and structural properties of glucocorticoid-induced TNF receptor ligand: Implications for function.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2R1F
 
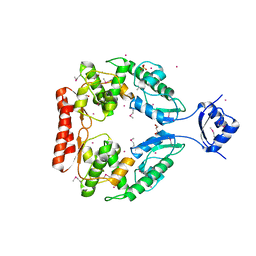 | | Crystal structure of predicted aminodeoxychorismate lyase from Escherichia coli | | Descriptor: | CADMIUM ION, GLYCEROL, Predicted aminodeoxychorismate lyase, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Rutter, M, Lau, C, Maletic, M, Smith, D, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Predicted Aminodeoxychorismate Lyase from Escherichia coli.
To be Published
|
|
2R30
 
 | |
