7EN4
 
 | | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase YUH1 | | Authors: | Okada, M, Tateishi, Y, Nojiri, E, Mikawa, T, Rajesh, S, Ogasawa, H, Ueda, T, Yagi, H, Kohno, T, Kigawa, T, Shimada, I, Guentert, P, Yutaka, I, Ikeya, T. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi-state structure determination and dynamics analysis elucidate a new ubiquitin-recognition mechanism of yeast ubiquitin C-terminal hydrolase.
To Be Published
|
|
2KM7
 
 | | Solution Structure of BamE, a component of the outer membrane protein assembly machinery in Escherichia coli | | Descriptor: | Small protein A | | Authors: | Knowles, T.J, Sridhar, P, Rajesh, S, Manoli, E, Henderson, I.R, Overduin, M. | | Deposit date: | 2009-07-24 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of BamE within the outer membrane and the beta-barrel assembly machine.
Embo Rep., 12, 2011
|
|
6R0X
 
 | | The extracellular domain of G6b-B in complex with Fab fragment and DP12 heparin oligosaccharide. | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Megakaryocyte and platelet inhibitory receptor G6b, antibody fab fragment heavy chain, ... | | Authors: | Ogg, D.J, McMiken, H.J, Howard, T.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Heparan sulfates are critical regulators of the inhibitory megakaryocyte-platelet receptor G6b-B.
Elife, 8, 2019
|
|
2MXC
 
 | | Solution structure of the full length sorting nexin 3 | | Descriptor: | Sorting nexin-3 | | Authors: | Lenoir, M.M.L, Rajesh, S.S.R, Gruenberg, J.J.G, Overduin, M.M.O, Kaur, J.J.K. | | Deposit date: | 2014-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of conserved phosphoinositide binding pocket regulates sorting nexin membrane targeting.
Nat Commun, 9, 2018
|
|
6T28
 
 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 19 (CS640) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[2,6-di(propan-2-yl)pyridin-4-yl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
6T29
 
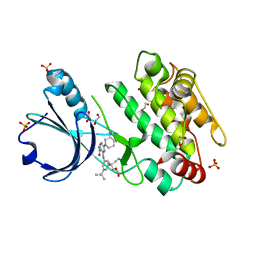 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) bound to compound 18 (CS587) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3~{S})-3-azanylpiperidin-1-yl]-4-[[3,5-bis(2-cyanopropan-2-yl)phenyl]amino]pyrimidine-5-carboxamide, Calcium/calmodulin-dependent protein kinase type 1D, ... | | Authors: | Kraemer, A, Sorrell, F, Butterworth, S, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-08 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Discovery of Highly Selective Inhibitors of Calmodulin-Dependent Kinases That Restore Insulin Sensitivity in the Diet-Induced Obesityin VivoMouse Model.
J.Med.Chem., 63, 2020
|
|
2OO5
 
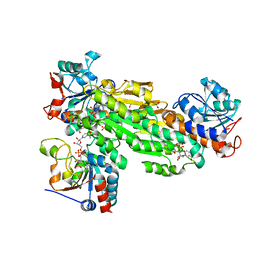 | | Structure of transhydrogenase (dI.H2NADH)2(dIII.NADP+)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
2OOR
 
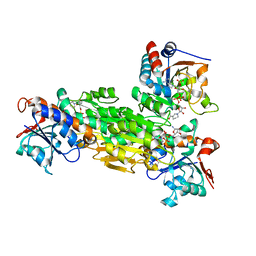 | | Structure of transhydrogenase (dI.NAD+)2(dIII.H2NADPH)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-26 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
