6WWX
 
 | |
6WV2
 
 | |
6WXA
 
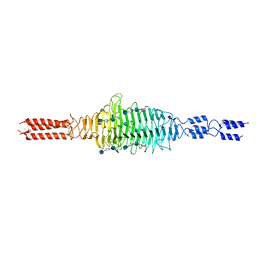
 | | Crystal structure of truncated Streptococcal bacteriophage hyaluronidase complexed with unsaturated hyaluronan hexa-saccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-10 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
6X3M
 
 | | Crystal structure of full-length Streptococcal bacteriophage hyaluronidase in complex with unsaturated hyaluronan octa-saccharides | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hyaluronoglucosaminidase | | Authors: | Deivanayagam, C, Schormann, N. | | Deposit date: | 2020-05-21 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Crystal Structure of Streptococcal Bacteriophage Hyaluronidase:
Presence of a Prokaryotic Collagen and Elucidation of Catalytic Mechanism
To Be Published
|
|
5T0O
 
 | | Crystal Structure of a membrane protein | | Descriptor: | CmeB | | Authors: | Su, C.-C, Yu, E.W. | | Deposit date: | 2016-08-16 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures and transport dynamics of a Campylobacter jejuni multidrug efflux pump.
Nat Commun, 8, 2017
|
|
5UI8
 
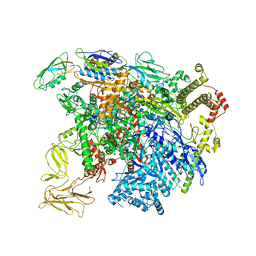 | | structure of sigmaN-holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Darst, S.A, Campbell, E.A. | | Deposit date: | 2017-01-13 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.76 Å) | | Cite: | Crystal structure of Aquifex aeolicus sigma (N) bound to promoter DNA and the structure of sigma (N)-holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4JYO
 
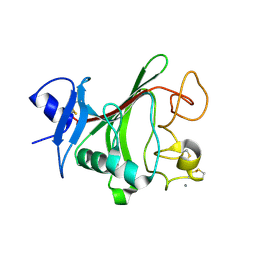 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, CALCIUM ION | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-03-31 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JZC
 
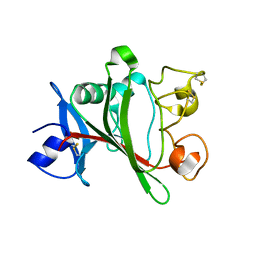 | | Angiopoietin-2 fibrinogen domain TAG mutant | | Descriptor: | Angiopoietin-2 | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-02 | | Release date: | 2013-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K0V
 
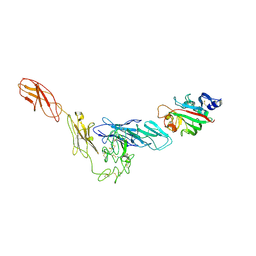 | | Structural basis for angiopoietin-1 mediated signaling initiation | | Descriptor: | Angiopoietin-1, TEK tyrosine kinase variant | | Authors: | Yu, X, Seegar, T.C.M, Dalton, A.C, Tzvetkova-Robev, D, Goldgur, Y, Nikolov, D.B, Barton, W.A. | | Deposit date: | 2013-04-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.51 Å) | | Cite: | Structural basis for angiopoietin-1-mediated signaling initiation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4DOP
 
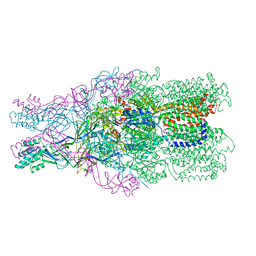 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, R mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-10 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
4DNT
 
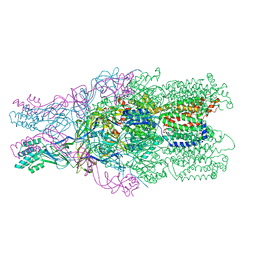 | | Crystal structure of the CusBA heavy-metal efflux complex from Escherichia coli, mutant | | Descriptor: | Cation efflux system protein CusA, Cation efflux system protein CusB | | Authors: | Su, C.-C, Long, F, Yu, E. | | Deposit date: | 2012-02-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Charged Amino Acids (R83, E567, D617, E625, R669, and K678) of CusA Are Required for Metal Ion Transport in the Cus Efflux System.
J.Mol.Biol., 422, 2012
|
|
4DOH
 
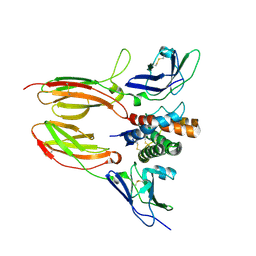 | | IL20/IL201/IL20R2 Ternary Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-20, Interleukin-20 receptor subunit alpha, ... | | Authors: | Logsdon, N.J, Walter, M.R. | | Deposit date: | 2012-02-09 | | Release date: | 2012-07-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OQS
 
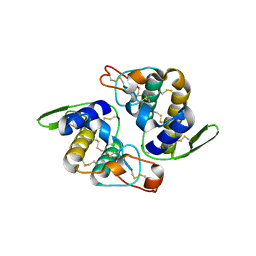 | | Crystal Structure of RV4/RV7 Complex | | Descriptor: | Phospholipase A2 RV-4, Phospholipase A2 RV-7 | | Authors: | Perbandt, M, Betzel, C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimeric neurotoxic complex viperotoxin F (RV-4/RV-7) from the venom of Vipera russelli formosensis at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6BMN
 
 | | Structure of human DHHC20 palmitoyltransferase, space group P63 | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ZINC ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6CAM
 
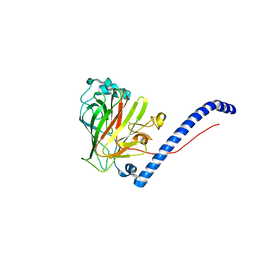 | |
6BMM
 
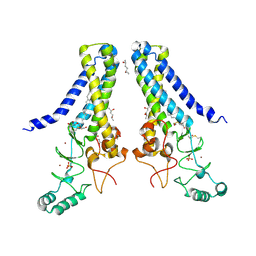 | | Structure of human DHHC20 palmitoyltransferase, space group P21 | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,5S)-hexane-2,5-diol, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
1Q3H
 
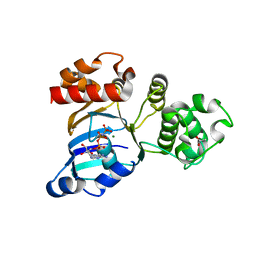 | | mouse CFTR NBD1 with AMP.PNP | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
5KHS
 
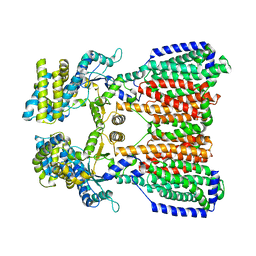 | |
5KHN
 
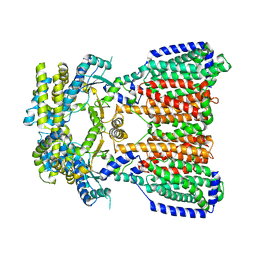 | |
5LQ3
 
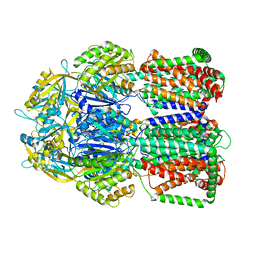 | |
1X94
 
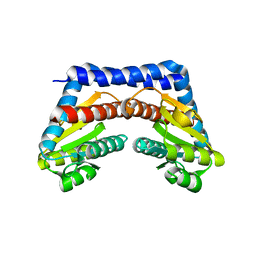 | |
5BSA
 
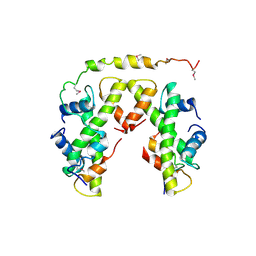 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.611 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5BS7
 
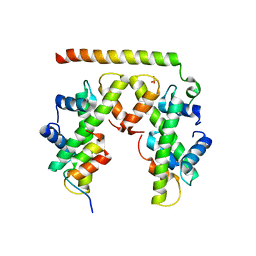 | | Structure of histone H3/H4 in complex with Spt2 | | Descriptor: | Histone H3.2, Histone H4, Protein SPT2 homolog, ... | | Authors: | Chen, S, Patel, D.J. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-function studies of histone H3/H4 tetramer maintenance during transcription by chaperone Spt2.
Genes Dev., 29, 2015
|
|
5D1W
 
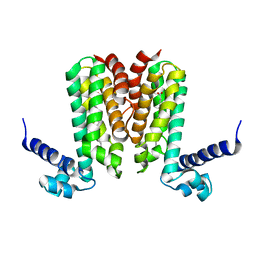 | | Crystal structure of Mycobacterium tuberculosis Rv3249c transcriptional regulator. | | Descriptor: | PALMITIC ACID, Rv3249c transcriptional regulator | | Authors: | Chou, T.-H, Delmar, J, Su, C.-C, Yu, E. | | Deposit date: | 2015-08-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Basis for the Regulation of the MmpL Transporters of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
