4U5I
 
 | |
4U5K
 
 | |
4RQZ
 
 | |
4R0D
 
 | | Crystal structure of a eukaryotic group II intron lariat | | Descriptor: | GROUP IIB INTRON LARIAT, IRIDIUM HEXAMMINE ION, LIGATED EXONS, ... | | Authors: | Robart, A.R, Chan, R.T, Peters, J.K, Rajashankar, K.R, Toor, N. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Crystal structure of a eukaryotic group II intron lariat.
Nature, 514, 2014
|
|
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
6J0L
 
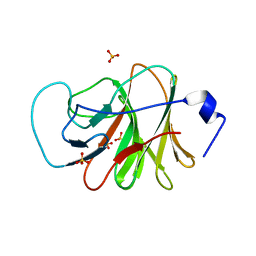 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with sulfate ion | | Descriptor: | Butyrophilin subfamily 3 member A3, SULFATE ION | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J8W
 
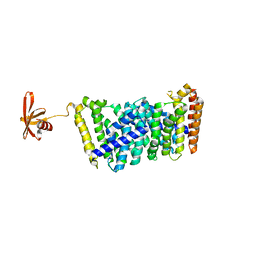 | | Structure of MOEN5-SSO7D fusion protein in complex with lig 1 | | Descriptor: | (2S)-3-dimethoxyphosphoryloxy-2-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6J0K
 
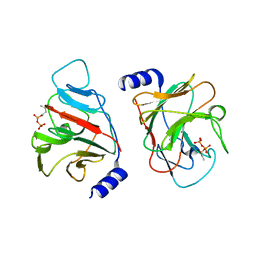 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
3DDW
 
 | | Crystal structure of glycogen phosphorylase complexed with an anthranilimide based inhibitor GSK055 | | Descriptor: | (2S)-{[(3-{[(2-chloro-6-methylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]amino}(phenyl)ethanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6J0G
 
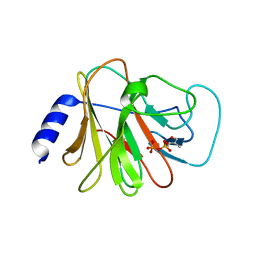 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
3DDS
 
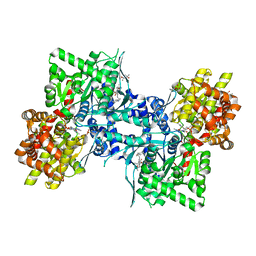 | |
6J03
 
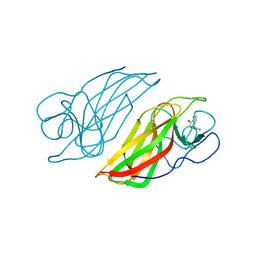 | | Crystal structure of a cyclase mutant in apo form | | Descriptor: | 12-epi-hapalindole U synthase, CALCIUM ION, amino({3-[(3S,8aS)-1,4-dioxooctahydropyrrolo[1,2-a]pyrazin-3-yl]propyl}amino)methaniminium | | Authors: | Tang, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the calcium dependence of Stig cyclases.
Rsc Adv, 9, 2019
|
|
3EQ2
 
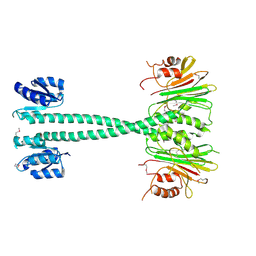 | |
6J06
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP-08 | | Descriptor: | (2E)-3-(hydroxymethyl)-4-(4-methylphenyl)but-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1, CALCIUM ION, ... | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6J8V
 
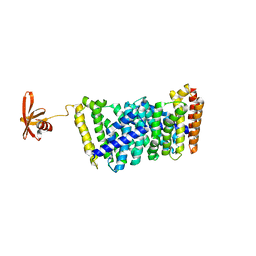 | | Structure of MOEN5-SSO7D fusion protein in complex with ligand 2 | | Descriptor: | FARNESYL, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6JJS
 
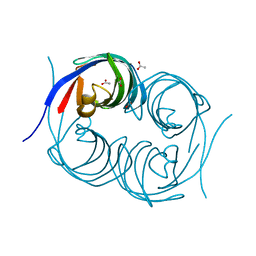 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
3CJF
 
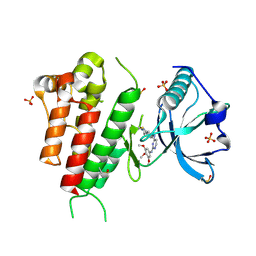 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
4O8J
 
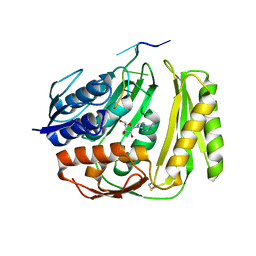 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii, in complex with rACAAA3'phosphate and adenine. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE, RNA, ... | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
6JJT
 
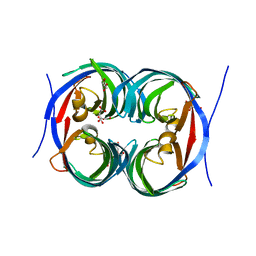 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
4O89
 
 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
3DNJ
 
 | | The structure of the Caulobacter crescentus ClpS protease adaptor protein in complex with a N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adapter protein clpS, MAGNESIUM ION, synthetic N-end rule peptide | | Authors: | Wang, K, Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2008-07-02 | | Release date: | 2008-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The molecular basis of N-end rule recognition.
Mol.Cell, 32, 2008
|
|
3EOD
 
 | |
6KQX
 
 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | Descriptor: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
4OQU
 
 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92)) | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, SAM-I/IV riboswitch | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
