8SGW
 
 | | Pendrin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-04-13 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
3K3F
 
 | |
3K3G
 
 | | Crystal Structure of the Urea Transporter from Desulfovibrio Vulgaris Bound to 1,3-dimethylurea | | Descriptor: | 1,3-dimethylurea, GOLD ION, Urea transporter | | Authors: | Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2009-10-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a bacterial homologue of the kidney urea transporter.
Nature, 462, 2009
|
|
3QNQ
 
 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
6D9Z
 
 | | Structure of CysZ, a sulfate permease from Pseudomonas Denitrificans | | Descriptor: | Sulfate transporter CysZ, octyl beta-D-glucopyranoside | | Authors: | Sanghai, Z.A, Clarke, O.B, Liu, Q, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-30 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4021318 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
6D79
 
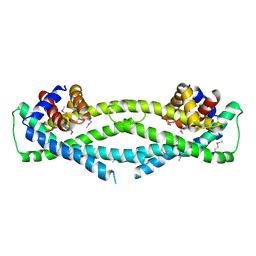 | | Structure of CysZ, a sulfate permease from Pseudomonas Fragi | | Descriptor: | Sulfate transporter CysZ | | Authors: | Sanghai, Z.A, Liu, Q, Clarke, O.B, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Mancia, F. | | Deposit date: | 2018-04-24 | | Release date: | 2018-05-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structure-based analysis of CysZ-mediated cellular uptake of sulfate.
Elife, 7, 2018
|
|
3PJZ
 
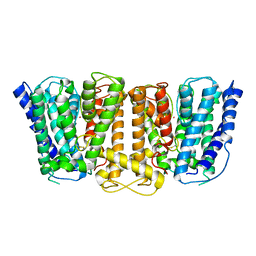 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
4EZC
 
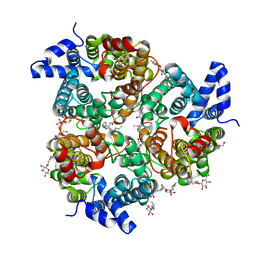 | | Crystal Structure of the UT-B Urea Transporter from Bos Taurus | | Descriptor: | OCTANOIC ACID (2-HYDROXY-1-HYDROXYMETHYL-HEPTADEC-3-ENYL)-AMIDE, Urea transporter 1, beta-D-glucopyranose, ... | | Authors: | Cao, Y, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2012-05-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and permeation mechanism of a mammalian urea transporter.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EZD
 
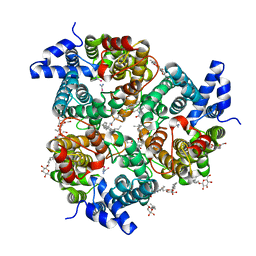 | | Crystal Structure of the UT-B Urea Transporter from Bos Taurus Bound to Selenourea | | Descriptor: | OCTANOIC ACID (2-HYDROXY-1-HYDROXYMETHYL-HEPTADEC-3-ENYL)-AMIDE, Urea transporter 1, beta-D-glucopyranose, ... | | Authors: | Cao, Y, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2012-05-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and permeation mechanism of a mammalian urea transporter.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
4N7X
 
 | |
6NQ8
 
 | |
6NQ9
 
 | |
6NQ7
 
 | |
6XWM
 
 | | Mechanism of substrate release in neurotransmitter:sodium symporters: the structure of LeuT in an inward-facing occluded conformation | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), PHENYLALANINE, SODIUM ION | | Authors: | Boesen, T, Nissen, P, Gotfryd, K, Loland, C.J, Gether, U. | | Deposit date: | 2020-01-24 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of LeuT in an inward-facing occluded conformation reveals mechanism of substrate release.
Nat Commun, 11, 2020
|
|
7DJC
 
 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DIX
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
