1Q5X
 
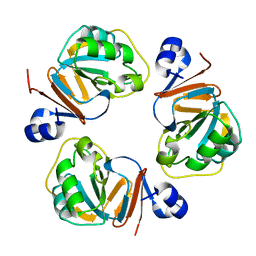 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
7Z6T
 
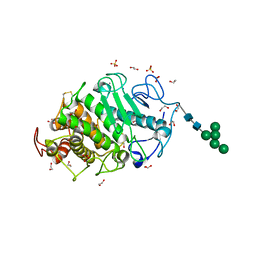 | | Aspergillus clavatus M36 protease without the propeptide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Extracellular metalloproteinase mep, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2022-03-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Aspergillus clavatus M36 protease without the propeptide
To Be Published
|
|
5CRC
 
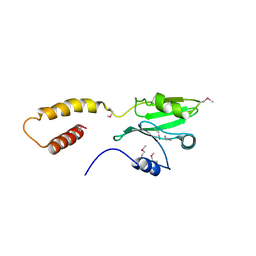 | | Structure of the SdeA DUB Domain | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Tan, Y, Paul, L.N, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CRB
 
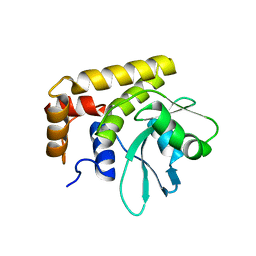 | | Crystal Structure of SdeA DUB | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CRA
 
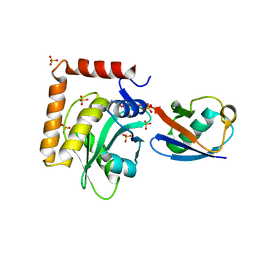 | | Structure of the SdeA DUB Domain | | Descriptor: | METHYL 4-AMINOBUTANOATE, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7QFY
 
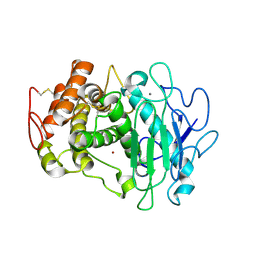 | | Fusarium oxysporum M36 protease without the propeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular metalloproteinase, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fusarium oxysporum M36 protease without the propeptide
To Be Published
|
|
7QP3
 
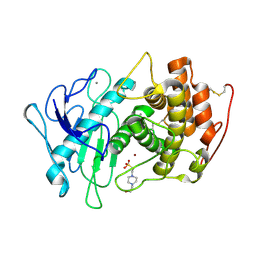 | | Pseudogymnoascus pannorum M36 protease without the propeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wilkens, C, Qiu, J, Meyer, A.S, Morth, J.P. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phaeosphaeria nodorum M36 protease without the propeptide
To Be Published
|
|
1I3A
 
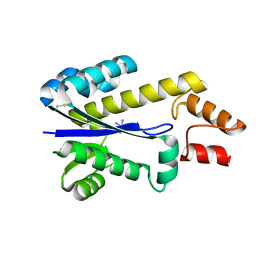 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS WITH COBALT HEXAMMINE CHLORIDE | | Descriptor: | COBALT HEXAMMINE(III), RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
1I39
 
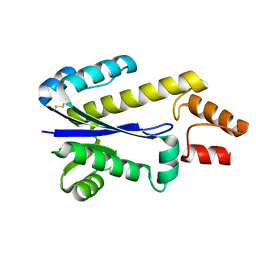 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
4TRG
 
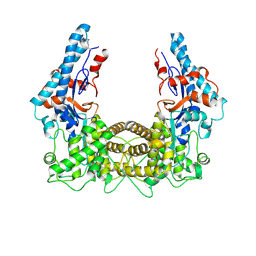 | | the SNL domain of SidC | | Descriptor: | MERCURY (II) ION, SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TRH
 
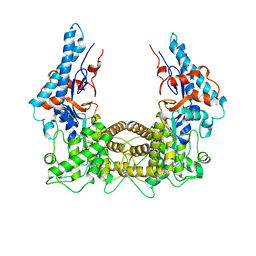 | | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling | | Descriptor: | SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1RXV
 
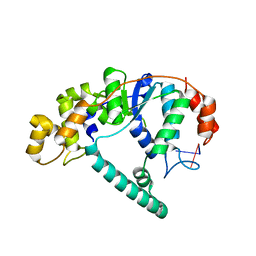 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXW
 
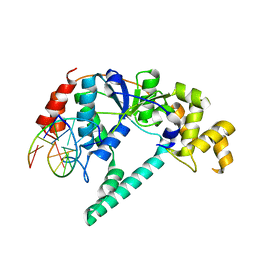 | | Crystal structure of A. fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*C*pG*pA*pT*pG*pC*pT*pA)-3', 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG)-3', Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXZ
 
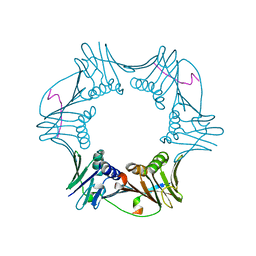 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RWZ
 
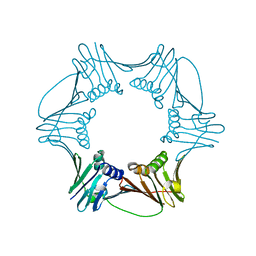 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) from A. fulgidus | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-17 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1RXM
 
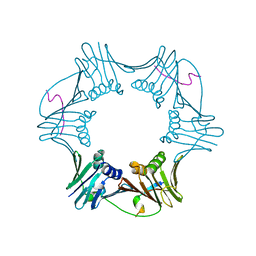 | | C-terminal region of FEN-1 bound to A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, consensus FEN-1 peptide | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3H09
 
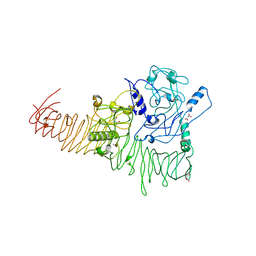 | | The structure of Haemophilus influenzae IgA1 protease | | Descriptor: | ACETATE ION, Immunoglobulin A1 protease, MALONIC ACID, ... | | Authors: | Johnson, T.A, Qiu, J, Plaut, A.G, Holyoak, T. | | Deposit date: | 2009-04-08 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Active-Site Gating Regulates Substrate Selectivity in a Chymotrypsin-Like Serine Protease The Structure of Haemophilus influenzae Immunoglobulin A1 Protease.
J.Mol.Biol., 389, 2009
|
|
8IVM
 
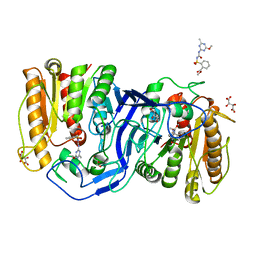 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVS
 
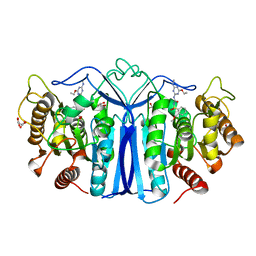 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8J7J
 
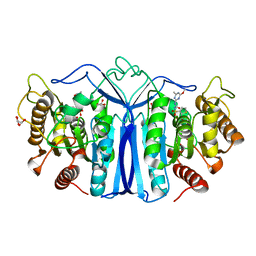 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW6
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVE
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVN
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IVT
 
 | | crystal structure of SulE mutant | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-28 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8IW3
 
 | | crystal structure of SulE mutant | | Descriptor: | 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, GLYCEROL, ... | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
