7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
2BCW
 
 | | Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L7/L12, Elongation factor G | | Authors: | Datta, P.P, Sharma, M.R, Qi, L, Frank, J, Agrawal, R.K. | | Deposit date: | 2005-10-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Interaction of the G' Domain of Elongation Factor G and the C-Terminal Domain of Ribosomal Protein L7/L12 during Translocation as Revealed by Cryo-EM.
Mol.Cell, 20, 2005
|
|
6BGJ
 
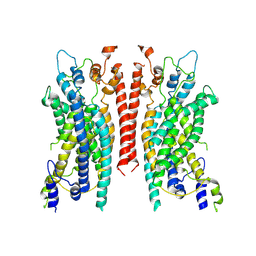 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
8Y3N
 
 | | The self-assembled nanotube of CPC46A/Q70C | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
8Y3T
 
 | | The self-assembled nanotube of CPC46A/Q70H | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
8Y3V
 
 | | The self-assembled nanotube of CPC46A/Q70V | | Descriptor: | Capsid protein | | Authors: | Yang, M, Rao, G, Li, L, Qi, L, Ma, C, Zhang, H, Gong, J, Wei, B, Zhang, X.E, Chen, G, Cao, S, Li, F. | | Deposit date: | 2024-01-29 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Transformation of a Viral Capsid from Nanocages to Nanotubes and Then to Hydrogels: Redirected Self-Assembly and Effects on Immunogenicity.
Acs Nano, 18, 2024
|
|
4RB1
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RAY
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Liu, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RAZ
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 holo-Fur | | Descriptor: | 1,2-ETHANEDIOL, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Wang, Q, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB3
 
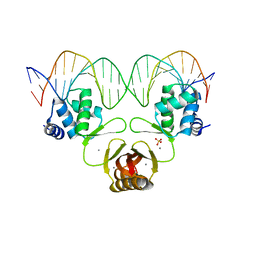 | |
4RB0
 
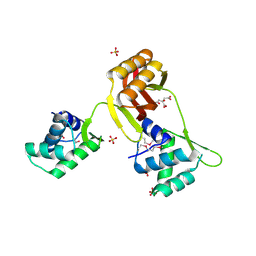 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 SeMet-Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB2
 
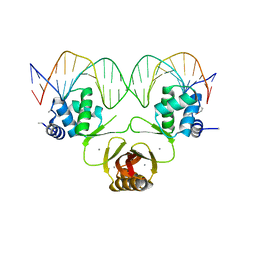 | |
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
9IV1
 
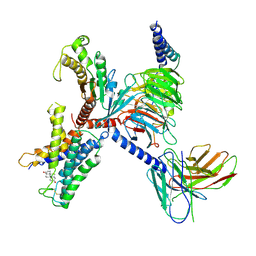 | | Identification, structure and agonist design of an androgen membrane receptor. | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y.Q, Yang, Z. | | Deposit date: | 2024-07-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Identification, structure, and agonist design of an androgen membrane receptor.
Cell, 188, 2025
|
|
9IV2
 
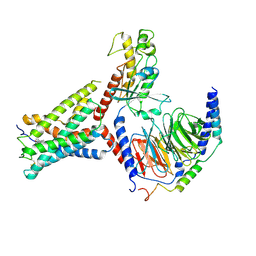 | | Identification, structure and agonist design of an androgen membrane receptor. | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Adhesion G-protein coupled receptor D1, Gs protein alpha subunit, ... | | Authors: | Ping, Y.Q, Yang, Z. | | Deposit date: | 2024-07-22 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Identification, structure, and agonist design of an androgen membrane receptor.
Cell, 188, 2025
|
|
7WNV
 
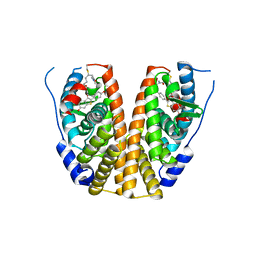 | | Crystal structure of mutant estrogen receptor alpha Y537S in complex with CO9 | | Descriptor: | (~{Z})-4-[2-[4-[[2-(4-hydroxyphenyl)-6-oxidanyl-1-benzothiophen-3-yl]oxy]phenoxy]ethylamino]-~{N},~{N}-dimethyl-but-2-enamide, Estrogen receptor | | Authors: | Xiao, Y, Lv, Y. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallography study and optimization of novel benzothiophene analogs as potent selective estrogen receptor covalent antagonists (SERCAs) with improved potency and safety profiles.
Bioorg.Chem., 141, 2023
|
|
7E4I
 
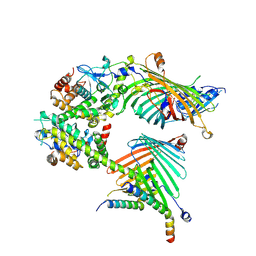 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40/Tom5/Tom6 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, Mitochondrial import receptor subunit TOM6, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
7E4H
 
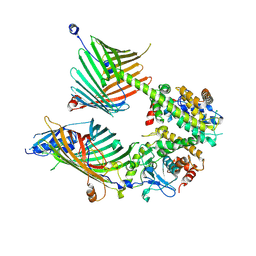 | | Cryo-EM structure of the yeast mitochondrial SAM-Tom40 complex at 3.0 angstrom | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Wang, Q, Guan, Z.Y, Qi, L.B, Yan, C.Y, Yin, P. | | Deposit date: | 2021-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Science, 373, 2021
|
|
5WWD
 
 | | Crystal structure of AtNUDX1 | | Descriptor: | AMMONIUM ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2016-12-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.386 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
5WY6
 
 | | Crystal structure of AtNUDX1 (E56A) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nudix hydrolase 1, ... | | Authors: | Liu, J, Guan, Z, Yan, L, Zou, T, Yin, P. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of Arabidopsis GPP-Bound NUDX1 for Noncanonical Monoterpene Biosynthesis.
Mol Plant, 11, 2018
|
|
6LSV
 
 | | Crystal structure of JOX2 in complex with 2OG, Fe, and JA | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Probable 2-oxoglutarate-dependent dioxygenase At5g05600, ... | | Authors: | Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure-guided analysis of Arabidopsis JASMONATE-INDUCED OXYGENASE (JOX) 2 reveals key residues for recognition of jasmonic acid substrate by plant JOXs.
Mol Plant, 14, 2021
|
|
7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
7EWQ
 
 | | Structure of Mumps virus nucleocapsid ring | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Su, X, Shen, Q, Shan, H. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-23 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
7EXA
 
 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
8X9S
 
 | | Identification, structure and agonist design of an androgen membrane receptor. | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Adhesion G-protein coupled receptor D1, Gs protein alpha subunit, ... | | Authors: | Ping, Y.Q, Yang, Z. | | Deposit date: | 2023-12-01 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Identification, structure, and agonist design of an androgen membrane receptor.
Cell, 188, 2025
|
|
