6T3N
 
 | |
6T3R
 
 | |
6T3K
 
 | |
8G7U
 
 | |
8G7T
 
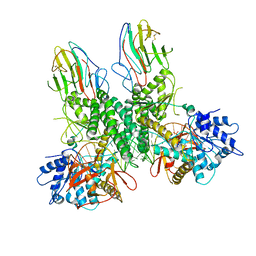 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
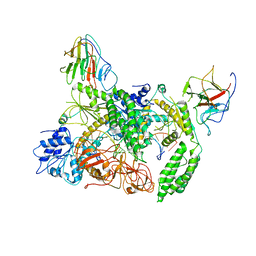 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
2YKG
 
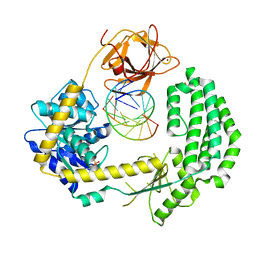 | | Structural insights into RNA recognition by RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
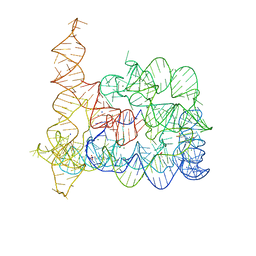
6T3S
 
 | |
1R2P
 
 | | Solution structure of domain 5 from the ai5(gamma) group II intron | | Descriptor: | 34-MER | | Authors: | Sigel, R.K.O, Sashital, D.G, Abramovitz, D.L, Palmer III, A.G, Butcher, S.E, Pyle, A.M. | | Deposit date: | 2003-09-29 | | Release date: | 2004-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif.
Nat.Struct.Mol.Biol., 11, 2004
|
|
7MK1
 
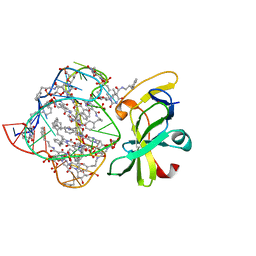 | | Structure of a protein-modified aptamer complex | | Descriptor: | Antiviral innate immune response receptor RIG-I, DNA (41-MER), MAGNESIUM ION, ... | | Authors: | Ren, X, Pyle, A.M. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolving A RIG-I Antagonist: A Modified DNA Aptamer Mimics Viral RNA.
J.Mol.Biol., 433, 2021
|
|
3BWP
 
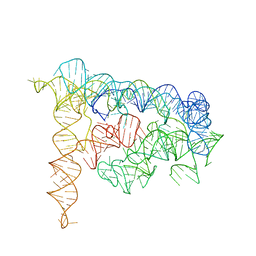 | | Crystal structure of a self-spliced group II intron | | Descriptor: | Group IIC intron, MAGNESIUM ION, POTASSIUM ION | | Authors: | Toor, N, Keating, K.S, Taylor, S.D, Pyle, A.M. | | Deposit date: | 2008-01-10 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a self-spliced group II intron
Science, 320, 2008
|
|
8DVR
 
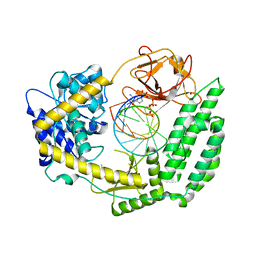 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVS
 
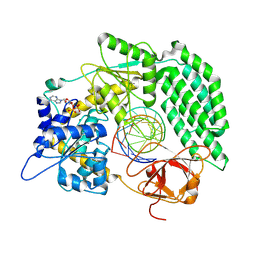 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVU
 
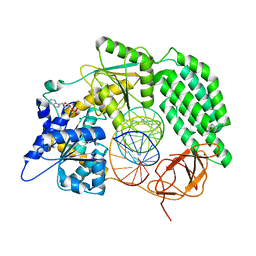 | |
5WDX
 
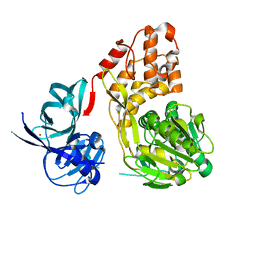 | |
4Y1O
 
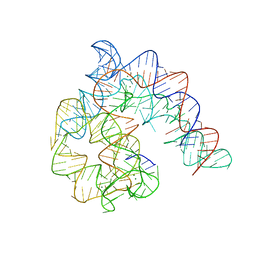 | | Oceanobacillus iheyensis group II intron domain 1 | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, group II intron, ... | | Authors: | Zhao, C, Rajashankar, K.R, Marcia, M, Pyle, A.M. | | Deposit date: | 2015-02-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of group II intron domain 1 reveals a template for RNA assembly.
Nat.Chem.Biol., 11, 2015
|
|
4Y1N
 
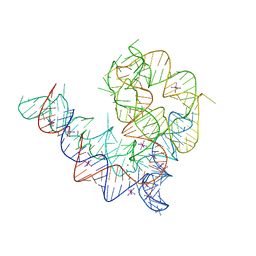 | | Oceanobacillus iheyensis group II intron domain 1 with iridium hexamine | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Zhao, C, Rajashankar, K.R, Marcia, M, Pyle, A.M. | | Deposit date: | 2015-02-08 | | Release date: | 2015-10-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of group II intron domain 1 reveals a template for RNA assembly.
Nat.Chem.Biol., 11, 2015
|
|
8SCZ
 
 | |
8SD0
 
 | |
8T2T
 
 | | Structure of a group II intron ribonucleoprotein in the post-ligation (post-2F) state | | Descriptor: | AMMONIUM ION, Group II intron reverse transcriptase/maturase, MAGNESIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2R
 
 | | Structure of a group II intron ribonucleoprotein in the pre-ligation (pre-2F) state | | Descriptor: | 5'exon, AMMONIUM ION, CALCIUM ION, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
8T2S
 
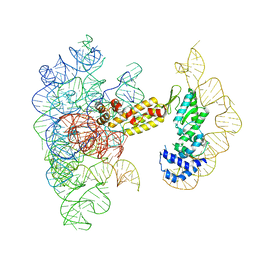 | | Structure of a group II intron ribonucleoprotein in the pre-branching (pre-1F) state | | Descriptor: | AMMONIUM ION, CALCIUM ION, Group II intron reverse transcriptase/maturase, ... | | Authors: | Xu, L, Liu, T, Chung, K, Pyle, A.M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into intron catalysis and dynamics during splicing.
Nature, 624, 2023
|
|
3EOG
 
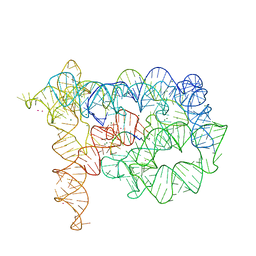 | | Co-crystallization showing exon recognition by a group II intron | | Descriptor: | 5'-R(*UP*UP*AP*UP*UP*A)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Rajashankar, K, Keating, K.S, Pyle, A.M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Structural basis for exon recognition by a group II intron.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3EOH
 
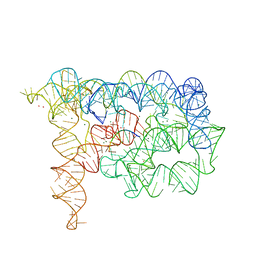 | | Refined group II intron structure | | Descriptor: | 5'-R(*UP*UP*AP*UP*UP*A)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Rajashankar, K, Keating, K.S, Pyle, A.M. | | Deposit date: | 2008-09-26 | | Release date: | 2008-10-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | Structural basis for exon recognition by a group II intron.
Nat.Struct.Mol.Biol., 15, 2008
|
|
7TNY
 
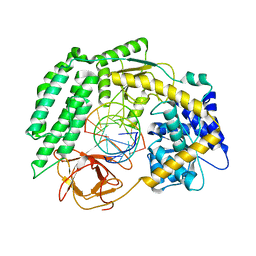 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
