6SWG
 
 | |
3OUV
 
 | |
5E12
 
 | |
5E10
 
 | |
5E0Y
 
 | |
5E0Z
 
 | |
4M6G
 
 | | Structure of the Mycobacterium tuberculosis peptidoglycan amidase Rv3717 in complex with L-Alanine-iso-D-Glutamine reaction product | | Descriptor: | ALANINE, D-alpha-glutamine, Peptidoglycan Amidase Rv3717, ... | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6I
 
 | | Structure of the reduced, Zn-bound form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717, ZINC ION | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.666 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4M6H
 
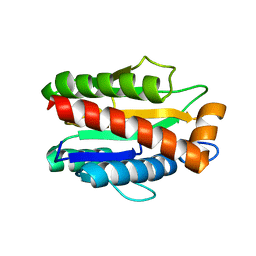 | | Structure of the reduced, metal-free form of Mycobacterium tuberculosis peptidoglycan amidase Rv3717 | | Descriptor: | Peptidoglycan Amidase Rv3717 | | Authors: | Prigozhin, D.M, Mavrici, D, Huizar, J.P, Vansell, H.J, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural and Biochemical Analyses of Mycobacterium tuberculosis N-Acetylmuramyl-L-alanine Amidase Rv3717 Point to a Role in Peptidoglycan Fragment Recycling.
J.Biol.Chem., 288, 2013
|
|
4PPR
 
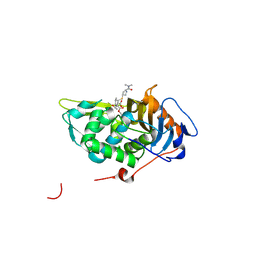 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | Authors: | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
4CGE
 
 | |
4ESQ
 
 | | Crystal structure of the extracellular domain of PknH from Mycobacterium tuberculosis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Serine/threonine protein kinase, TERBIUM(III) ION | | Authors: | Cavazos, A, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-04-23 | | Release date: | 2012-07-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the sensor domain of Mycobacterium tuberculosis PknH receptor kinase reveals a conserved binding cleft.
J.Mol.Biol., 422, 2012
|
|
4P0M
 
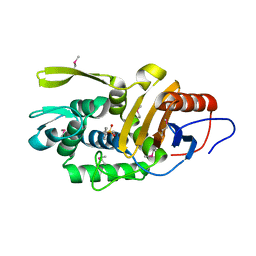 | | Crystal structure of an evolved putative penicillin-binding protein homolog, Rv2911, from Mycobacterium tuberculosis | | Descriptor: | D-alanyl-D-alanine carboxypeptidase | | Authors: | Krieger, I, Yu, M, Bursey, E, Hung, L.-W, Terwilliger, T.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subfamily-Specific Adaptations in the Structures of Two Penicillin-Binding Proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
6TL1
 
 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
6VPT
 
 | |
4G1H
 
 | |
4G1J
 
 | | Sortase C1 of GBS Pilus Island 1 | | Descriptor: | Sortase family protein | | Authors: | Cozzi, R, Prigozhin, D.M, Alber, T. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for group B streptococcus pilus 1 sortases C regulation and specificity.
Plos One, 7, 2012
|
|
4N8N
 
 | | Crystal structure of Mycobacterial FtsX extracellular domain | | Descriptor: | Cell division protein FtsX, POTASSIUM ION | | Authors: | Mavrici, D. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N8O
 
 | |
5T5V
 
 | | LIPOXYGENASE-1 (SOYBEAN) AT 293K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
5TR0
 
 | | Lipoxygenase-1 (soybean) L754A mutant at 293K | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
5TQP
 
 | | LIPOXYGENASE-1 (SOYBEAN) I553G MUTANT AT 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-Deuterium Exchange of Lipoxygenase Uncovers a Relationship between Distal, Solvent Exposed Protein Motions and the Thermal Activation Barrier for Catalytic Proton-Coupled Electron Tunneling.
ACS Cent Sci, 3, 2017
|
|
5TQO
 
 | | Lipoxygenase-1 (soybean) L546A/L754A mutant at 300K | | Descriptor: | FE (III) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
5TQN
 
 | | Lipoxygenase-1 (soybean) L546A mutant at 293K | | Descriptor: | FE (II) ION, Seed linoleate 13S-lipoxygenase-1 | | Authors: | Poss, E.M, Fraser, J.S, Gee, C. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Characterization of a Disabled Double Mutant of Soybean Lipoxygenase: The "Undoing" of Precise Substrate Positioning Relative to Metal Cofactor and an Identified Dynamical Network.
J.Am.Chem.Soc., 141, 2019
|
|
