4JKW
 
 | |
4JRY
 
 | | Crystal Structure of SB47 TCR-HLA B*3505-LPEP complex | | Descriptor: | Beta-2-microglobulin, MAGNESIUM ION, MHC class I antigen, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule.
J.Biol.Chem., 288, 2013
|
|
4JRX
 
 | | Crystal Structure of CA5 TCR-HLA B*3505-LPEP complex | | Descriptor: | Beta-2-microglobulin, CA5 TCR alpha chain, CA5 TCR beta chain, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Highly divergent T-cell receptor binding modes underlie specific recognition of a bulged viral peptide bound to a human leukocyte antigen class I molecule.
J.Biol.Chem., 288, 2013
|
|
4K55
 
 | |
5T6Z
 
 | | KIR3DL1 in complex with HLA-B*57:01-TW10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-THR-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6Y
 
 | | HLA-B*57:01 presenting TSTFEDVKILAF | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Decapeptide: THR-SER-THR-PHE-GLU-ASP-VAL-LYS-ILE-LEU-ALA-PHE, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T70
 
 | | KIR3DL1 in complex with HLA-B*57:01 presenting TSNLQEQIGW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-ASN-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6W
 
 | | HLA-B*57:01 presenting SSTRGISQLW | | Descriptor: | Beta-2-microglobulin, Decapeptide: SER-SER-THR-ARG-GLY-ILE-SER-GLN-LEU-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6X
 
 | | HLA-B*57:01 presenting TSTTSVASSW | | Descriptor: | Beta-2-microglobulin, Decapeptide: THR-SER-THR-THR-SER-VAL-ALA-SER-SER-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5WKH
 
 | | D30 TCR in complex with HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS3 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WKF
 
 | | D30 TCR in complex with HLA-A*11:01-GTS1 | | Descriptor: | Beta-2-microglobulin, D30 TCR beta chain, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WJN
 
 | | Crystal Structure of HLA-A*11:01-GTS3 | | Descriptor: | Beta-2-microglobulin, GTS3 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WLG
 
 | | Crystal Structure of H-2Db with the GAP501 peptide (SQL) | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GAP50 peptide, ... | | Authors: | Gras, S, Farenc, C, Josephs, T, Rossjohn, J. | | Deposit date: | 2017-07-26 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A T Cell Receptor Locus Harbors a Malaria-Specific Immune Response Gene.
Immunity, 47, 2017
|
|
5WJL
 
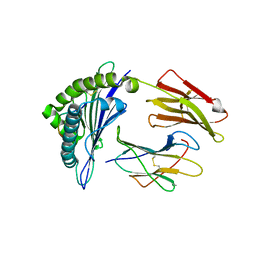 | | Crystal Structure of HLA-A*11:01 with GTS1 peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GTS1 peptide, ... | | Authors: | Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-23 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Nat. Immunol., 18, 2017
|
|
5WLI
 
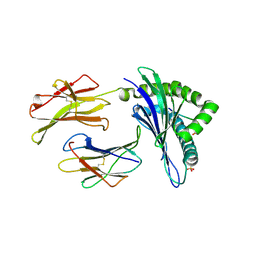 | | Crystal Structure of H-2Db with the GAP501 peptide (SQL) | | Descriptor: | Beta-2-microglobulin, GAP50 peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Farenc, C, Josephs, T, Rossjohn, J. | | Deposit date: | 2017-07-27 | | Release date: | 2017-11-15 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A T Cell Receptor Locus Harbors a Malaria-Specific Immune Response Gene.
Immunity, 47, 2017
|
|
2MG9
 
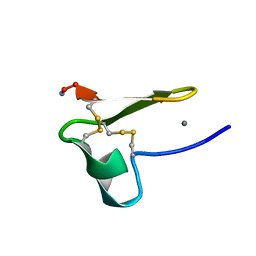 | | Truncated EGF-A | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor | | Authors: | Schroeder, C.I, Rosengren, K. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Method: | SOLUTION NMR | | Cite: | Design and Synthesis of Truncated EGF-A Peptides that Restore LDL-R Recycling in the Presence of PCSK9 In Vitro.
Chem.Biol., 21, 2014
|
|
6W0Z
 
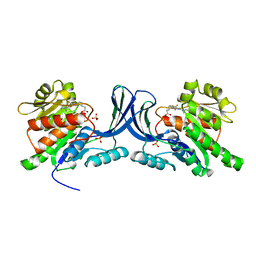 | |
6W0X
 
 | |
6W0W
 
 | | Structure of KHK in complex with compound 3 | | Descriptor: | 6-[(3~{R},4~{S})-3,4-bis(oxidanyl)pyrrolidin-1-yl]-2-[(2~{S},3~{R})-2-methyl-3-oxidanyl-azetidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, Ketohexokinase, SULFATE ION | | Authors: | Jasti, J. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of PF-06835919: A Potent Inhibitor of Ketohexokinase (KHK) for the Treatment of Metabolic Disorders Driven by the Overconsumption of Fructose.
J.Med.Chem., 63, 2020
|
|
6W0Y
 
 | |
6W0N
 
 | | Structure of KHK in complex with compound 2 | | Descriptor: | 6-[(3~{S},4~{R})-3,4-bis(oxidanyl)pyrrolidin-1-yl]-2-[(3~{S})-3-methyl-3-oxidanyl-pyrrolidin-1-yl]-4-(trifluoromethyl)pyridine-3-carbonitrile, CITRIC ACID, Ketohexokinase, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-03-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of PF-06835919: A Potent Inhibitor of Ketohexokinase (KHK) for the Treatment of Metabolic Disorders Driven by the Overconsumption of Fructose.
J.Med.Chem., 63, 2020
|
|
