6UPP
 
 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UV6
 
 | | AtmM with bound rebeccamycin analogue | | Descriptor: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
6V04
 
 | | DynU16 crystal structure, a putative protein in the dynemicin biosynthetic locus | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Alvarado, S.K, Miller, M.D, Bhardwaj, M, Thorson, J.S, Van Lanen, S.G, Phillips Jr, G.N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-04-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of DynU16, a START/Bet v1-like protein involved in dynemicin biosynthesis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
6VZX
 
 | | Structure of a Covalently Captured Collagen Triple Helix using Lysine-Glutamate Pairs | | Descriptor: | collagen mimetic peptide | | Authors: | Miller, M.D, Hulgan, S.A, Xu, W, Kosgei, A.J, Phillips Jr, G.N, Hartgerink, J.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Covalent Capture of Collagen Triple Helices Using Lysine-Aspartate and Lysine-Glutamate Pairs.
Biomacromolecules, 21, 2020
|
|
1MOA
 
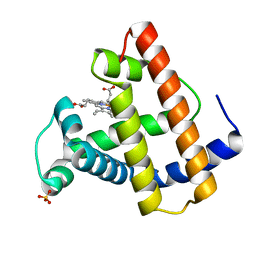 | |
4GLQ
 
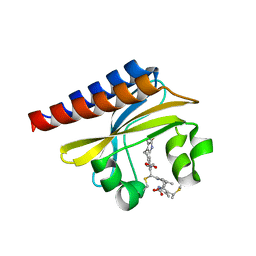 | | Crystal Structure of the blue-light absorbing form of the Thermosynechococcus elongatus PixJ GAF-domain | | Descriptor: | Methyl-accepting chemotaxis protein, Phycoviolobilin, blue light-absorbing form | | Authors: | Burgie, E.S, Walker, J.M, Phillips Jr, G.N, Vierstra, R.D. | | Deposit date: | 2012-08-14 | | Release date: | 2013-01-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | A Photo-Labile Thioether Linkage to Phycoviolobilin Provides the Foundation for the Blue/Green Photocycles in DXCF-Cyanobacteriochromes.
Structure, 21, 2013
|
|
6CKY
 
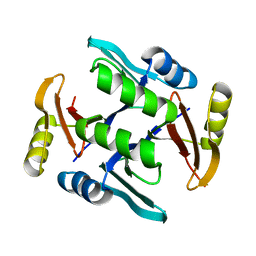 | | Crystal structure of UcmS2 | | Descriptor: | Glyoxalase | | Authors: | Chang, C.Y, Chang, C, Annaval, T, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of UcmS2
To Be Published
|
|
6D2V
 
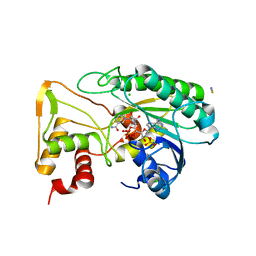 | | Apo Structure of TerB, an NADP Dependent Oxidoreductase in the Terfestatin Biosynthesis Pathway | | Descriptor: | CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, THIOCYANATE ION, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6D34
 
 | | Apo Crystal Structure of TerC, a Terfestatin Biosynthesis Enzyme | | Descriptor: | ISOPROPYL ALCOHOL, TerC | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | Descriptor: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DRU
 
 | | Xylosidase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Glycosyl hydrolases family 31 family protein, ... | | Authors: | Cao, H, Xu, W, Betancourt, M, Walton, J.D, Brumm, P, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of alpha-Xylosidase fromAspergillus nigerin Complex with a Hydrolyzed Xyloglucan Product and New Insights in Accurately Predicting Substrate Specificities of GH31 Family Glycosidases.
Acs Sustain Chem Eng, 8, 2020
|
|
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2014-04-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
1MLU
 
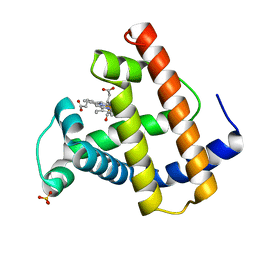 | |
4Z5Q
 
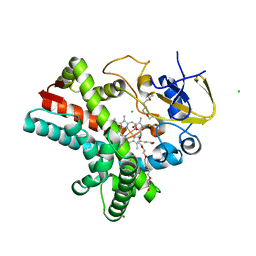 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4ZAS
 
 | | Crystal structure of sugar aminotransferase CalS13 from Micromonospora echinospora | | Descriptor: | CalS13, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, F, Singh, S, Miller, M.D, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure characterization of sugar aminotransferases CalS13 and WecE provides the basis for a unifying structural model for stereochemical outcome.
To Be Published
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
5CQF
 
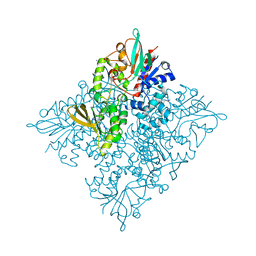 | | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae | | Descriptor: | IODIDE ION, L-lysine 6-monooxygenase | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Weerth, R.S, Cao, H, Yennamalli, R, Phillips Jr, G.N, Thomas, M.G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of L-lysine 6-monooxygenase from Pseudomonas syringae
To Be Published
|
|
5DU2
 
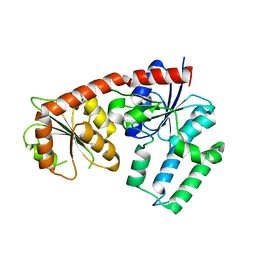 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
1MOB
 
 | |
1MLN
 
 | |
1MLQ
 
 | |
