1BL8
 
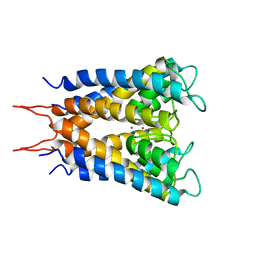 | | POTASSIUM CHANNEL (KCSA) FROM STREPTOMYCES LIVIDANS | | Descriptor: | POTASSIUM ION, PROTEIN (POTASSIUM CHANNEL PROTEIN) | | Authors: | Doyle, D.A, Cabral, J.M, Pfuetzner, R.A, Kuo, A, Gulbis, J.M, Cohen, S.L, Chait, B.T, Mackinnon, R. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the potassium channel: molecular basis of K+ conduction and selectivity.
Science, 280, 1998
|
|
1YJ7
 
 | | Crystal structure of enteropathogenic E.coli (EPEC) type III secretion system protein EscJ | | Descriptor: | GLYCEROL, PHOSPHATE ION, escJ | | Authors: | Yip, C.K, Kimbrough, T.G, Felise, H.B, Vuckovic, M, Thomas, N.A, Pfuetzner, R.A, Frey, E.A, Finlay, B.B, Miller, S.I, Strynadka, N.C.J. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the molecular platform for type III secretion system assembly.
Nature, 435, 2005
|
|
1F00
 
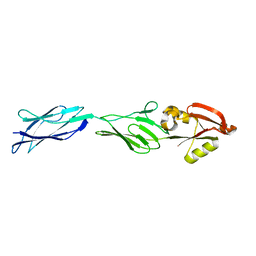 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | Descriptor: | INTIMIN | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F02
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF INTIMIN IN COMPLEX WITH TRANSLOCATED INTIMIN RECEPTOR (TIR) INTIMIN-BINDING DOMAIN | | Descriptor: | INTIMIN, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-14 | | Release date: | 2000-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1JHF
 
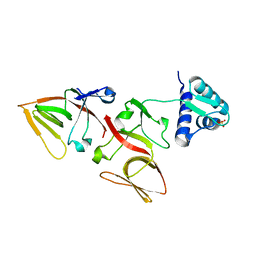 | | LEXA G85D MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1K3E
 
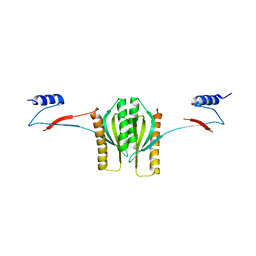 | | Type III secretion chaperone CesT | | Descriptor: | CesT | | Authors: | Luo, Y, Bertero, M, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3S
 
 | | Type III Secretion Chaperone SigE | | Descriptor: | PHOSPHATE ION, SigE | | Authors: | Bertero, M.G, Luo, Y, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-28 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1JHH
 
 | | LEXA S119A MUTANT | | Descriptor: | LEXA REPRESSOR, SULFATE ION | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHC
 
 | | LEXA S119A C-TERMINAL TRYPTIC FRAGMENT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, Strynadka, N.C.J. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JHE
 
 | | LEXA L89P Q92W E152A K156A MUTANT | | Descriptor: | LEXA REPRESSOR | | Authors: | Luo, Y, Pfuetzner, R.A, Mosimann, S, Little, J.W, J Strynadka, N.C. | | Deposit date: | 2001-06-27 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of LexA: a conformational switch for regulation of self-cleavage.
Cell(Cambridge,Mass.), 106, 2001
|
|
1VHI
 
 | | EPSTEIN BARR VIRUS NUCLEAR ANTIGEN-1 DNA-BINDING DOMAIN, RESIDUES 470-607 | | Descriptor: | EPSTEIN BARR VIRUS NUCLEAR ANTIGEN-1 | | Authors: | Bochkarev, A, Barwell, J, Pfuetzner, R, Furey, W, Edwards, A, Frappier, L. | | Deposit date: | 1996-10-05 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein EBNA 1.
Cell(Cambridge,Mass.), 83, 1995
|
|
8CZI
 
 | |
6MDM
 
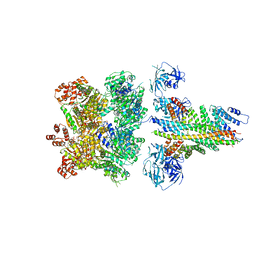 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MDP
 
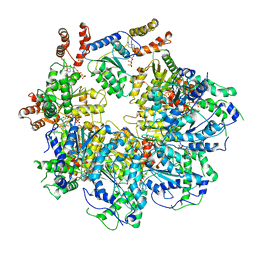 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MDO
 
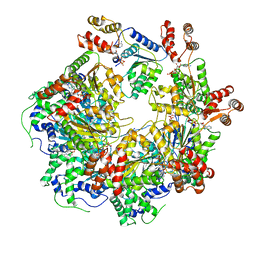 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
6MDN
 
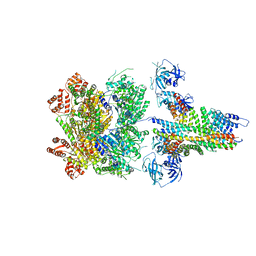 | | The 20S supercomplex engaging the SNAP-25 N-terminus (class 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-soluble NSF attachment protein, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
1JMC
 
 | | SINGLE STRANDED DNA-BINDING DOMAIN OF HUMAN REPLICATION PROTEIN A BOUND TO SINGLE STRANDED DNA, RPA70 SUBUNIT, RESIDUES 183-420 | | Descriptor: | DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*C)-3'), PROTEIN (REPLICATION PROTEIN A (RPA)) | | Authors: | Bochkarev, A, Pfuetzner, R, Edwards, A, Frappier, L. | | Deposit date: | 1996-11-11 | | Release date: | 1997-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA.
Nature, 385, 1997
|
|
4WY4
 
 | | Crystal structure of autophagic SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Zhao, M, Brunger, A.T. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes.
Nature, 520, 2015
|
|
1B3T
 
 | | EBNA-1 NUCLEAR PROTEIN/DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*AP*GP*CP*AP*TP*AP*TP*GP*CP*TP*TP*CP*CP*C)-3'), PROTEIN (NUCLEAR PROTEIN EBNA1) | | Authors: | Bochkarev, A, Bochkareva, E, Edwards, A, Frappier, L. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1.
J.Mol.Biol., 284, 1998
|
|
1Y9L
 
 | | The X-ray structure of an secretion system protein | | Descriptor: | ACETATE ION, Lipoprotein mxiM, UNDECANE | | Authors: | Lario, P.I, Pfuetzer, R.A, Frey, E.A, Creagh, L, Haynes, C, Maurelli, A.T, Strynadka, N.C. | | Deposit date: | 2004-12-15 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and biochemical analysis of a secretin pilot protein.
Embo J., 24, 2005
|
|
4G1I
 
 | |
4G08
 
 | |
6PNP
 
 | | Crystal structure of the splice insert-free neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
6PNQ
 
 | | Crystal structure of the SS2A splice insert-containing neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
4G2S
 
 | |
