2YX2
 
 | | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution | | Descriptor: | Hyaluronidase, phage associated | | Authors: | Mishra, P, Prem Kumar, R, Bhakuni, V, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of cloned trimeric hyluranidase from streptococcus pyogenes at 2.8 A resolution
To be Published
|
|
2G32
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ4
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ6
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ5
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2GQ7
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, GLYCEROL, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), ... | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-20 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3GVN
 
 | | The 1.2 Angstroem crystal structure of an E.coli tRNASer acceptor stem microhelix reveals two magnesium binding sites | | Descriptor: | 5'-R(*CP*CP*UP*CP*AP*CP*C)-3', 5'-R(*GP*GP*UP*GP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Eichert, A, Furste, J.P, Schreiber, A, Perbandt, M, Betzel, C, Erdmann, V.A, Forster, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2A crystal structure of an E. coli tRNASer)acceptor stem microhelix reveals two magnesium binding sites.
Biochem.Biophys.Res.Commun., 386, 2009
|
|
1P7W
 
 | | Crystal structure of the complex of Proteinase K with a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Perbandt, M, Chandra, V, Banumathi, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the complex of Proteinase K with heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ser-Ala at atomic resolution
To be published
|
|
1P7V
 
 | | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution | | Descriptor: | CALCIUM ION, NITRATE ION, inhibitor peptide, ... | | Authors: | Bilgrami, S, Kaur, P, Chandra, V, Banumathi, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structure of a complex formed between Proteinase K and a designed heptapeptide inhibitor Pro-Ala-Pro-Phe-Ala-Ala-Ala at atomic resolution
To be published
|
|
1Q5T
 
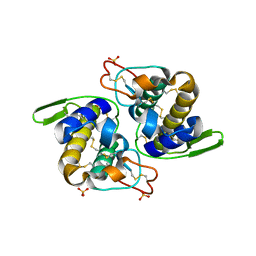 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
2MLL
 
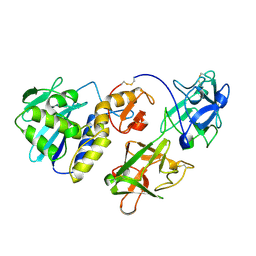 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-16 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
2G58
 
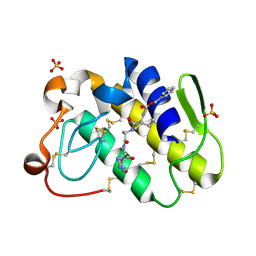 | | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution | | Descriptor: | (PHQ)IARS, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Prem Kumar, R, Singh, N, Somvanshi, R.K, Ethayathulla, A.S, Dey, S, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal structure of a complex of phospholipase A2 with a designed peptide inhibitor Dehydro-Ile-Ala-Arg-Ser at 0.98 A resolution
To be Published
|
|
2O1L
 
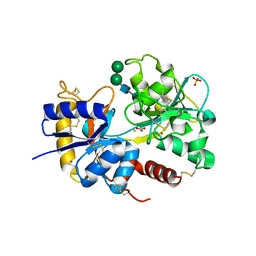 | | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Sharma, S, Perbandt, M, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.97 A resolution
To be Published
|
|
2NWJ
 
 | | Structure of the complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.75 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Sharma, S, Perbandt, M, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2006-11-15 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the complex of C-terminal lobe of bovine lactoferrin with disaccharide at 1.75 A resolution
To be Published
|
|
2OYF
 
 | | Crystal Structure of the complex of phospholipase A2 with indole acetic acid at 1.2 A resolution | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, ACETIC ACID, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the complex of phospholipase A2 with indole acetic acid at 1.2 A resolution
To be Published
|
|
2Q1P
 
 | | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution | | Descriptor: | N-PROPANOL, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Phospholipase A2 complex with propanol at 1.5 A resolution
To be Published
|
|
7B83
 
 | | Structure of SARS-CoV-2 Main Protease bound to pyrithione zinc | | Descriptor: | 3C-like proteinase, 9-oxa-7-thia-1-azonia-8$l^{2}-zincabicyclo[4.3.0]nona-1,3,5-triene, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
3UOU
 
 | | Crystal structure of the Kunitz-type protease inhibitor ShPI-1 Lys13Leu mutant in complex with pancreatic elastase | | Descriptor: | Chymotrypsin-like elastase family member 1, GLYCEROL, Kunitz-type proteinase inhibitor SHPI-1, ... | | Authors: | Garcia-Fernandez, R, Perbandt, M, Rehders, D, Gonzalez-Gonzalez, Y, Chavez, M.A, Betzel, C, Redecke, L. | | Deposit date: | 2011-11-17 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional Structure of a Kunitz-type Inhibitor in Complex with an Elastase-like Enzyme.
J.Biol.Chem., 290, 2015
|
|
3ZVA
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 75 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVG
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 98 | | Descriptor: | 3C PROTEASE, N-(tert-butoxycarbonyl)-O-tert-butyl-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZV8
 
 | |
3ZV9
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 74 | | Descriptor: | 3C PROTEASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVC
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 82 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
3ZVD
 
 | | 3C protease of Enterovirus 68 complexed with Michael receptor inhibitor 83 | | Descriptor: | 3C PROTEASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Perbandt, M, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-07-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3C Protease of Enterovirus 68: Structure-Based Design of Michael Acceptor Inhibitors and Their Broad-Spectrum Antiviral Effects Against Picornaviruses.
J.Virol., 87, 2013
|
|
