4V94
 
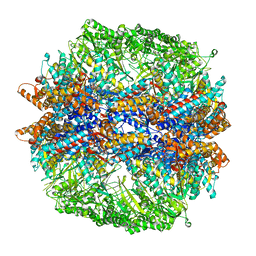 | | Molecular architecture of the eukaryotic chaperonin TRiC/CCT derived by a combination of chemical crosslinking and mass-spectrometry, XL-MS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Leitner, A, Joachimiak, L.A, Bracher, A, Walzthoeni, T, Chen, B, Monkemeyer, L, Pechmann, S, Holmes, S, Cong, Y, Ma, B, Ludtke, S, Chiu, W, Hartl, F.U, Aebersold, R, Frydman, J. | | Deposit date: | 2012-01-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Architecture of the Eukaryotic Chaperonin TRiC/CCT.
Structure, 20, 2012
|
|
5ER1
 
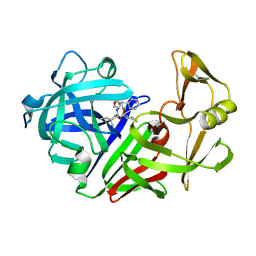 | |
3SLS
 
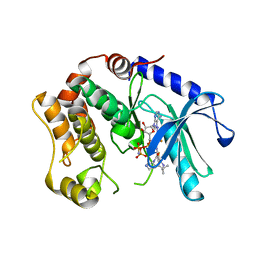 | | Crystal Structure of human MEK-1 kinase in complex with UCB1353770 and AMPPNP | | Descriptor: | 2-[(2-fluoro-4-iodophenyl)amino]-5,5-dimethyl-8-oxo-N-[(3R)-piperidin-3-yl]-5,6,7,8-tetrahydro-4H-thieno[2,3-c]azepine-3-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Meier, C, Ceska, T.A. | | Deposit date: | 2011-06-25 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering human MEK-1 for structural studies: A case study of combinatorial domain hunting.
J.Struct.Biol., 177, 2012
|
|
6QI9
 
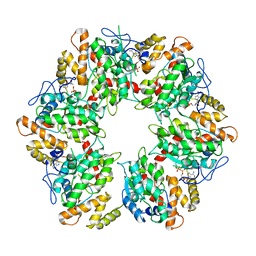 | | Truncated human R2TP complex, structure 4 (ADP-empty) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
4V8R
 
 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Kalisman, N, Schroder, G.F, Levitt, M. | | Deposit date: | 2012-03-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
1ENT
 
 | |
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
2J90
 
 | | Crystal structure of human ZIP kinase in complex with a tetracyclic pyridone inhibitor (Pyridone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, CHLORIDE ION, ... | | Authors: | Turnbull, A.P, Berridge, G, Fedorov, O, Pike, A.C.W, Savitsky, P, Eswaran, J, Papagrigoriou, E, Ugochukwa, E, von Delft, F, Gileadi, O, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2J7T
 
 | | Crystal structure of human serine threonine kinase-10 bound to SU11274 | | Descriptor: | (3Z)-N-(3-CHLOROPHENYL)-3-({3,5-DIMETHYL-4-[(4-METHYLPIPERAZIN-1-YL)CARBONYL]-1H-PYRROL-2-YL}METHYLENE)-N-METHYL-2-OXOINDOLINE-5-SULFONAMIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Das, S, Debreczeni, J, Sobott, F, Watt, S, Savitsky, P, Eswaran, J, Turnbull, A.P, Papagrigoriou, E, Ugochukwa, E, Gorrec, F, Umeano, C.C, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2J51
 
 | | Crystal structure of Human STE20-like kinase bound to 5-Amino-3-((4-(aminosulfonyl)phenyl)amino) -N-(2,6-difluorophenyl)-1H-1,2,4-triazole- 1-carbothioamide | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, STE20-LIKE SERINE/THREONINE-PROTEIN KINASE, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Turnbull, A.P, Debreczeni, J.E, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2JFM
 
 | | CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE (UNLIGANDED FORM) | | Descriptor: | 1,2-ETHANEDIOL, STE20-LIKE SERINE-THREONINE KINASE | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
2JFL
 
 | | CRYSTAL STRUCTURE OF HUMAN STE20-LIKE KINASE (DIPHOSPHORYLATED FORM) BOUND TO 5- AMINO-3-((4-(AMINOSULFONYL)PHENYL)AMINO)-N-(2,6- DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, CHLORIDE ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-02-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
1HFO
 
 | |
4ER2
 
 | | The active site of aspartic proteinases | | Descriptor: | ENDOTHIAPEPSIN, PEPSTATIN, SULFATE ION | | Authors: | Bailey, D, Veerapandian, B, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4ER1
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN, N-[(1R,2R,4R)-1-(cyclohexylmethyl)-2-hydroxy-6-methyl-4-{[(2R)-2-methylbutyl]carbamoyl}heptyl]-3-(1H-imidazol-3-ium-4-yl)-N~2~-[3-naphthalen-1-yl-2-(naphthalen-1-ylmethyl)propanoyl]-L-alaninamide | | Authors: | Quail, J.W, Cooper, J.B, Szelke, M, Blundell, T.L. | | Deposit date: | 1990-10-14 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The active site of aspartic proteinases
FEBS Lett., 174, 1984
|
|
4CGU
 
 | | Full length Tah1 bound to yeast PIH1 and HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, PROTEIN INTERACTING WITH HSP90 1, ... | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CHH
 
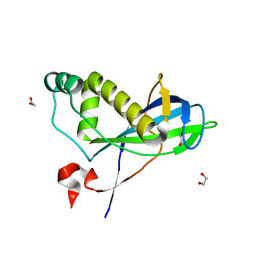 | | N-terminal domain of yeast PIH1p | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN INTERACTING WITH HSP90 1 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CV4
 
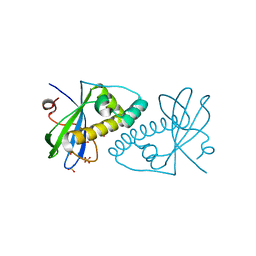 | | PIH N-terminal domain | | Descriptor: | COBALT (II) ION, PIH1 DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CKT
 
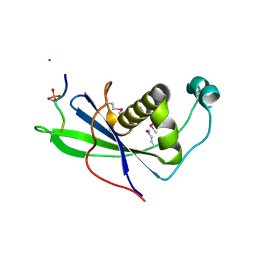 | | PIH1 N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-14 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CSE
 
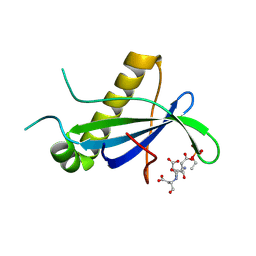 | | PIH N-terminal domain | | Descriptor: | PIH1 DOMAIN-CONTAINING PROTEIN 1, TELOMERE LENGTH REGULATION PROTEIN TEL2 HOMOLOG | | Authors: | Morgan, R.M, Roe, S.M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGV
 
 | | First TPR of Spaghetti (RPAP3) bound to HSP90 peptide SRMEEVD | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA, RNA POLYMERASE II-ASSOCIATED PROTEIN 3 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Basis for Phosphorylation-Dependent Recruitment of Tel2 to Hsp90 by Pih1.
Structure, 22, 2014
|
|
4CGQ
 
 | | Full length Tah1 bound to HSP90 peptide SRMEEVD | | Descriptor: | CHLORIDE ION, HEAT SHOCK PROTEIN HSP 90-ALPHA, TPR REPEAT-CONTAINING PROTEIN ASSOCIATED WITH HSP90 | | Authors: | Roe, S.M, Pal, M. | | Deposit date: | 2013-11-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tah1 Helix-Swap Dimerization Prevents Mixed Hsp90 Co-Chaperone Complexes
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4CGW
 
 | |
