3UIJ
 
 | |
3OWI
 
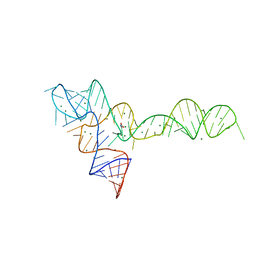 | |
3D1B
 
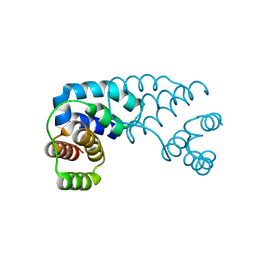 | | Tetragonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
3OXD
 
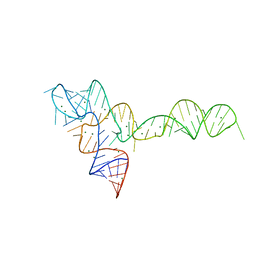 | |
3UIH
 
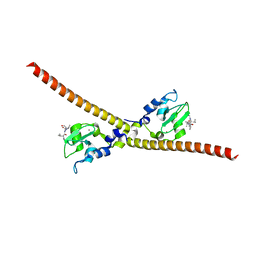 | |
3UIG
 
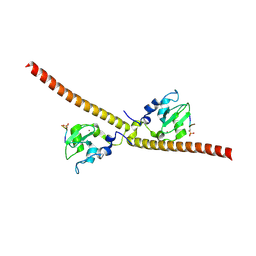 | |
3UII
 
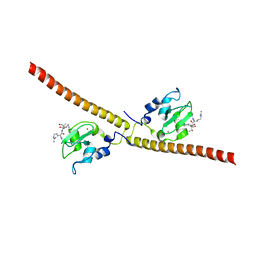 | |
3OWW
 
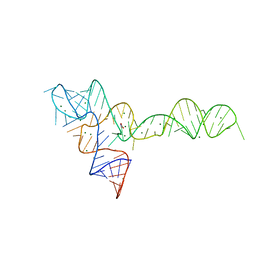 | |
3OXE
 
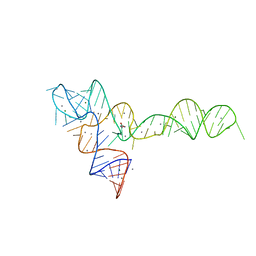 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OWZ
 
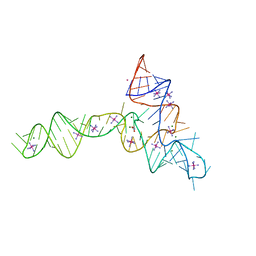 | | Crystal structure of glycine riboswitch, soaked in Iridium | | Descriptor: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXB
 
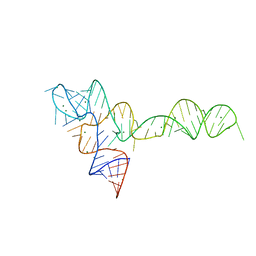 | |
3OXM
 
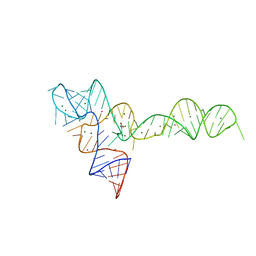 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3OXJ
 
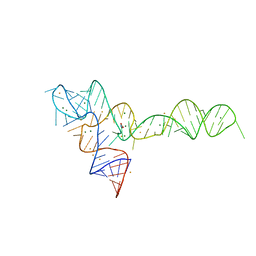 | | crystal structure of glycine riboswitch, soaked in Ba2+ | | Descriptor: | BARIUM ION, GLYCINE, MAGNESIUM ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3P04
 
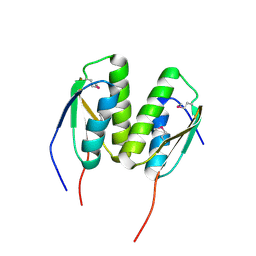 | | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8 | | Descriptor: | Uncharacterized BCR | | Authors: | Vorobiev, S, Lew, S, Seetharaman, J, Hamilton, H, Xiao, R, Patel, D.J, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-27 | | Release date: | 2010-10-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the BCR protein from Corynebacterium glutamicum. Northeast Structural Genomics Consortium Target CgR8.
To be Published
|
|
3DLH
 
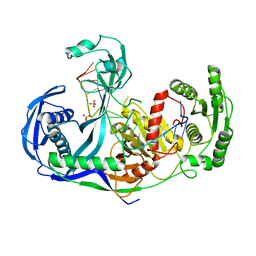 | | Crystal structure of the guide-strand-containing Argonaute protein silencing complex | | Descriptor: | ACETIC ACID, Argonaute, DNA (5'-D(DTP*DGP*DAP*DGP*DGP*DTP*DAP*DGP*DTP*DAP*DGP*DGP*DTP*DTP*DGP*DTP*DAP*DTP*DAP*DGP*DT)-3'), ... | | Authors: | Wang, Y, Sheng, G, Patel, D.J. | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the guide-strand-containing argonaute silencing complex.
Nature, 456, 2008
|
|
3DLB
 
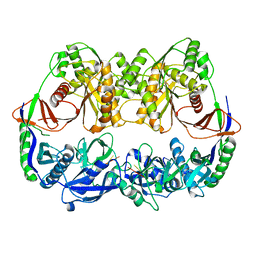 | |
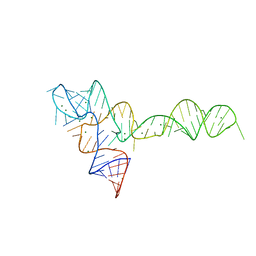
3OX0
 
 | |
3PT6
 
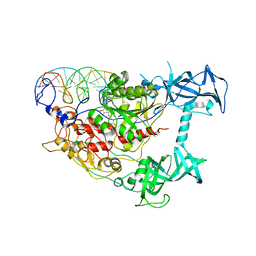 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3Q0D
 
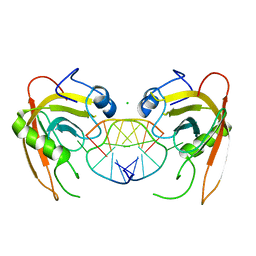 | | Crystal structure of SUVH5 SRA- hemi methylated CG DNA complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*AP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*(5CM)P*GP*TP*CP*AP*G)-3'), ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3704 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PTA
 
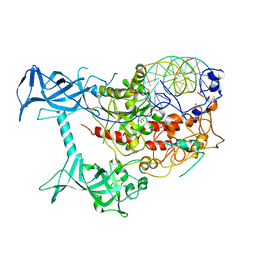 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3VRS
 
 | | Crystal structure of fluoride riboswitch, soaked in Mn2+ | | Descriptor: | FLUORIDE ION, Fluoride riboswitch, MANGANESE (II) ION, ... | | Authors: | Ren, A.M, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch.
Nature, 486, 2012
|
|
3Q0B
 
 | |
3Q0F
 
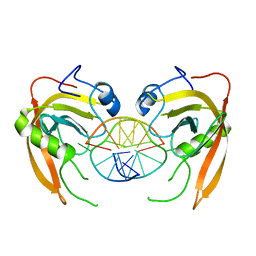 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3Q0C
 
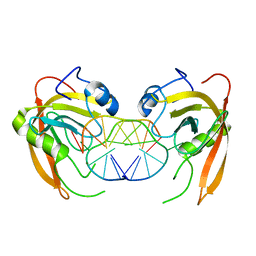 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6567 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3PT9
 
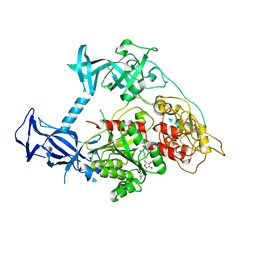 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
