1SH0
 
 | | Crystal Structure of Norwalk Virus Polymerase (Triclinic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SH3
 
 | | Crystal Structure of Norwalk Virus Polymerase (MgSO4 crystal form) | | Descriptor: | MAGNESIUM ION, RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1SH2
 
 | | Crystal Structure of Norwalk Virus Polymerase (Metal-free, Centered Orthorhombic) | | Descriptor: | RNA Polymerase | | Authors: | Ng, K.K, Pendas-Franco, N, Rojo, J, Boga, J.A, Machin, A, Alonso, J.M, Parra, F. | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft.
J.Biol.Chem., 279, 2004
|
|
1KHW
 
 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
1KHV
 
 | | Crystal Structure of Rabbit Hemorrhagic Disease Virus RNA-dependent RNA polymerase complexed with Lu3+ | | Descriptor: | LUTETIUM (III) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ng, K.K, Cherney, M.M, Vazquez, A.L, Machin, A, Alonso, J.M, Parra, F, James, M.N. | | Deposit date: | 2001-12-01 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase.
J.Biol.Chem., 277, 2002
|
|
4QPX
 
 | | NV polymerase post-incorporation-like complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Polyprotein, ... | | Authors: | Zamyatkin, D.F, Parra, F, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2014-06-25 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of a backtracked state reveals conformational changes similar to the state following nucleotide incorporation in human norovirus polymerase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3H5X
 
 | | Crystal Structure of 2'-amino-2'-deoxy-cytidine-5'-triphosphate bound to Norovirus GII RNA polymerase | | Descriptor: | 2'-amino-2'-deoxycytidine 5'-(tetrahydrogen triphosphate), 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
3H5Y
 
 | | Norovirus polymerase+primer/template+CTP complex at 6 mM MnCl2 | | Descriptor: | 5'-R(*UP*GP*CP*CP*CP*GP*GP*G)-3', 5'-R(P*UP*GP*CP*CP*CP*GP*GP*GP*C)-3', CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Zamyatkin, D.F, Parra, F, Machin, A, Grochulski, P, Ng, K.K.S. | | Deposit date: | 2009-04-22 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of 2'-amino-2'-deoxycytidine-5'-triphosphate to norovirus polymerase induces rearrangement of the active site.
J.Mol.Biol., 390, 2009
|
|
2VIM
 
 | | X-ray structure of Fasciola hepatica thioredoxin | | Descriptor: | THIOREDOXIN | | Authors: | Line, K, Isupov, M.N, Garcia-Rodriguez, E, Maggioli, G, Parra, F, Littlechild, J.A. | | Deposit date: | 2007-12-05 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Fasciola Hepatica Thioredoxin: High Resolution Structure Reveals Two Oxidation States.
Mol.Biochem.Parasitol., 161, 2008
|
|
3BSO
 
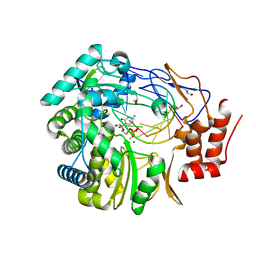 | |
3BSN
 
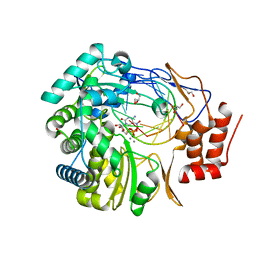 | |
8CYL
 
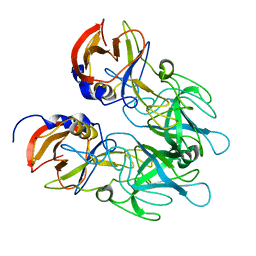 | | Ast89 P domain | | Descriptor: | Capsid protein VP60 | | Authors: | Leuthold, M, Kilic, T, Hansman, G. | | Deposit date: | 2022-05-23 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structural Basis for Rabbit Hemorrhagic Disease Virus Antibody Specificity.
J.Virol., 96, 2022
|
|
8CZ5
 
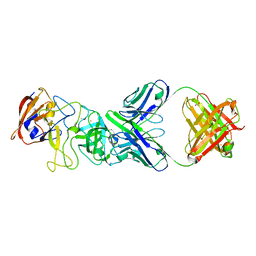 | | N11 P domain 2D9 Fab P complex | | Descriptor: | ACETATE ION, CADMIUM ION, Fab 2D9 Heavy Chain, ... | | Authors: | Leuthold, M, Kilic, T, Hansman, G. | | Deposit date: | 2022-05-24 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for Rabbit Hemorrhagic Disease Virus Antibody Specificity.
J.Virol., 96, 2022
|
|
