8DPW
 
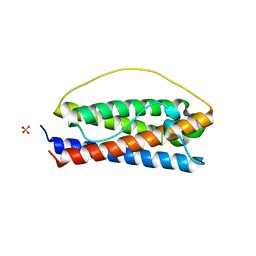 | | The structure of Interleukin-11 Mutein | | Descriptor: | Interleukin-11, SULFATE ION | | Authors: | Metcalfe, R.D, Griffin, M.D.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the interleukin 11 signalling complex reveal gp130 dynamics and the inhibitory mechanism of a cytokine variant.
Nat Commun, 14, 2023
|
|
8DPV
 
 | |
7ROU
 
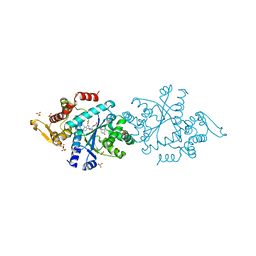 | | Structure of human tyrosyl tRNA synthetase in complex with ML901-Tyr | | Descriptor: | SULFATE ION, Tyrosine--tRNA ligase, cytoplasmic, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
7ROT
 
 | | Plasmodium falciparum tyrosyl-tRNA synthetase, S234C mutant, in complex with ML901-Tyr | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine--tRNA ligase, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
7ROS
 
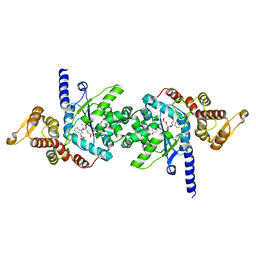 | | Plasmodium falciparum tyrosyl-tRNA synthetase in complex with ML901-Tyr | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine--tRNA ligase, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
7ROR
 
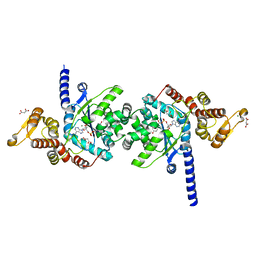 | | Plasmodium falciparum tyrosyl-tRNA synthetase in complex with tyrosine-AMP | | Descriptor: | 5'-O-[(S)-{[(2S)-2-amino-3-(4-hydroxyphenyl)propanoyl]oxy}(hydroxy)phosphoryl]adenosine, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Metcalfe, R.D, Xie, S.C, Morton, C.J, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy.
Science, 376, 2022
|
|
2FMA
 
 | |
3VFL
 
 | |
7LXV
 
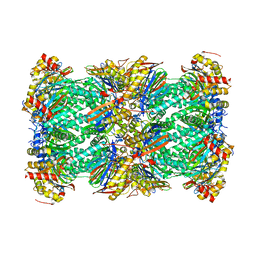 | | Structure of human 20S proteasome with bound MPI-5 | | Descriptor: | N-[(1R)-2-([1,1'-biphenyl]-4-yl)-1-boronoethyl]-1-methyl-L-prolinamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXT
 
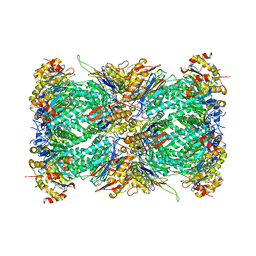 | | Structure of Plasmodium falciparum 20S proteasome with bound bortezomib | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Morton, C.J, Metcalfe, R.D, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXU
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound MPI-5 | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Xie, S.C, Liu, B, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EZV
 
 | |
6O4O
 
 | | The structure of human interleukin 11 | | Descriptor: | CHLORIDE ION, Interleukin-11, SULFATE ION | | Authors: | Metcalfe, R.D, Griffin, M.D.W. | | Deposit date: | 2019-02-28 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of the extracellular domains of human interleukin 11 alpha receptor reveals mechanisms of cytokine engagement.
J.Biol.Chem., 295, 2020
|
|
6CT2
 
 | | MYST histone acetyltransferase KAT6A/B in complex with WM-1119 | | Descriptor: | 3-fluoro-N'-[(2-fluorophenyl)sulfonyl]-5-(pyridin-2-yl)benzohydrazide, Histone acetyltransferase KAT8, MAGNESIUM ION, ... | | Authors: | Ren, B, Peat, T.S. | | Deposit date: | 2018-03-22 | | Release date: | 2018-08-01 | | Last modified: | 2018-08-22 | | Method: | X-RAY DIFFRACTION (2.128 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
3LE0
 
 | | Lectin Domain of Lectinolysin complexed with Glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-13 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LEI
 
 | | Lectin Domain of Lectinolysin complexed with Fucose | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LEK
 
 | | Lectin Domain of Lectinolysin complexed with Lewis B Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-15 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LEG
 
 | | Lectin Domain of Lectinolysin complexed with Lewis Y Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
6DFK
 
 | | Crystal structure of the 11S subunit of the Plasmodium falciparum proteasome, PA28 | | Descriptor: | SULFATE ION, Subunit of proteaseome activator complex,putative | | Authors: | Xie, S.C, Metcalfe, R.D, Gillett, D.L, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2018-05-15 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
3DLW
 
 | | Antichymotrypsin | | Descriptor: | Alpha-1-antichymotrypsin | | Authors: | Feil, S.C. | | Deposit date: | 2008-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and characterization of a misfolded monomeric serpin formed at physiological temperature
J.Mol.Biol., 403, 2010
|
|
3EIN
 
 | | Delta class GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1-1 | | Authors: | Feil, S.C. | | Deposit date: | 2008-09-17 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.126 Å) | | Cite: | Probing insect detoxification systems
To be Published
|
|
3SL4
 
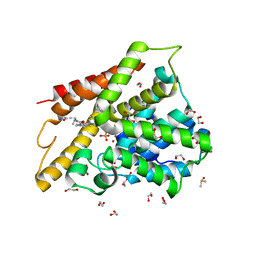 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL6
 
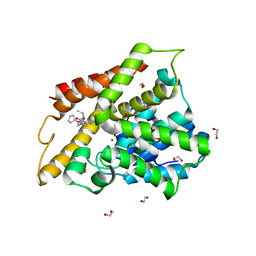 | | Crystal structure of the catalytic domain of PDE4D2 with compound 12c | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL8
 
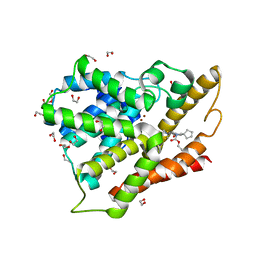 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL3
 
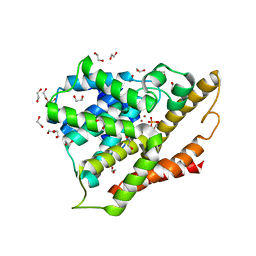 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
