7TC8
 
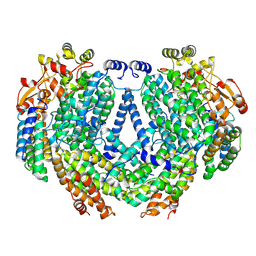 | | Cryo-EM structure of methane monooxygenase hydroxylase (by graphene) | | Descriptor: | FE (III) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Cho, U.S, Kim, B.C. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-25 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Batch Production of High-Quality Graphene Grids for Cryo-EM: Cryo-EM Structure of Methylococcus capsulatus Soluble Methane Monooxygenase Hydroxylase.
Acs Nano, 17, 2023
|
|
5KUT
 
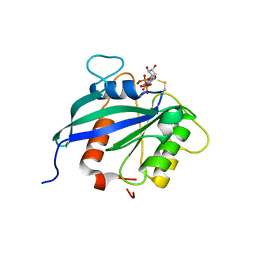 | | hMiro2 C-terminal GTPase domain, GDP-bound | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mitochondrial Rho GTPase 2 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSY
 
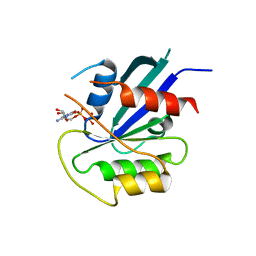 | | hMiro1 C-domain GDP Complex P41212 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSZ
 
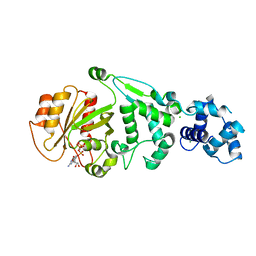 | | hMiro EF hand and cGTPase domains in the GMPPCP-bound state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mitochondrial Rho GTPase 1, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-10 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KTY
 
 | | hMiro EF hand and cGTPase domains, GDP and Ca2+ bound state | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.522 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSP
 
 | | hMiro1 C-domain GDP Complex C2221 Crystal Form | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1 | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KSO
 
 | | hMiro1 C-domain GDP-Pi Complex P3121 Crystal Form | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial Rho GTPase 1, PHOSPHATE ION | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5KU1
 
 | | hMiro1 EF hand and cGTPase domains in the GDP-bound state | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Rice, S.E, Freymann, D.M. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural insights into Parkin substrate lysine targeting from minimal Miro substrates.
Sci Rep, 6, 2016
|
|
5D6C
 
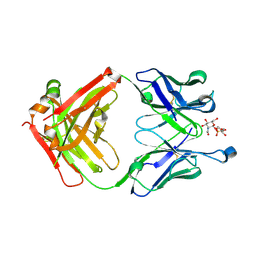 | | Structure of 4497 Fab bound to synthetic wall teichoic acid fragment | | Descriptor: | 4-O-[2-acetamido-2-deoxy-beta-D-glucopyranosyl]-5-O-phosphono-D-ribitol, 4497 antibody IgG1 (VH and CH1), 4497 antibody IgK (VL and CL), ... | | Authors: | Lupardus, P.J, Fong, R. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Novel antibody-antibiotic conjugate eliminates intracellular S. aureus.
Nature, 527, 2015
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | Descriptor: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | Deposit date: | 2019-11-02 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
6K8W
 
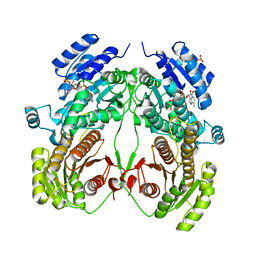 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
 | |
4BFM
 
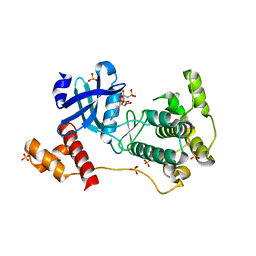 | | The crystal structure of mouse PK38 | | Descriptor: | MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Yoo, J.H, Cho, Y.S, Park, S.M, Cho, H.S. | | Deposit date: | 2013-03-21 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structures of the Kinase Domain and Uba Domain of Mpk38 Suggest the Activation Mechanism for Kinase Activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5DSL
 
 | |
5DSK
 
 | |
5DSQ
 
 | |
5DSM
 
 | |
5DSR
 
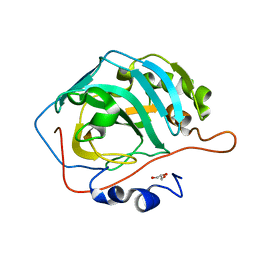 | |
5DSP
 
 | |
5DSO
 
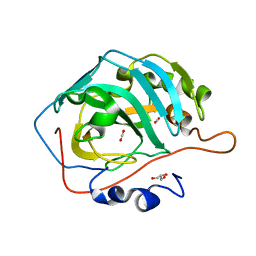 | |
6VFF
 
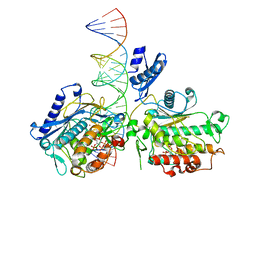 | | Dimer of Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from human GLI1 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*UP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Thuy-boun, A.S, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-01-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition.
Nucleic Acids Res., 48, 2020
|
|
6L4O
 
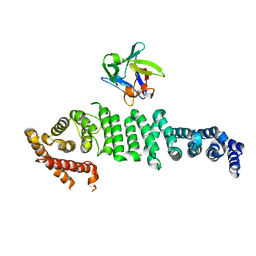 | | Crystal structure of API5-FGF2 complex | | Descriptor: | Apoptosis inhibitor 5, Fibroblast growth factor 2 | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of mRNA export through API5 and nuclear FGF2 interaction.
Nucleic Acids Res., 48, 2020
|
|
6P9L
 
 | |
6P9K
 
 | |
6P9M
 
 | |
