2A2L
 
 | |
8P33
 
 | | BB0238 from Borrelia burgdorferi | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
8P32
 
 | | BB0238 from Borrelia burgdorferi, Se-Met data for Leu240Met mutant | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
3BOV
 
 | | Crystal structure of the receptor binding domain of mouse PD-L2 | | Descriptor: | FORMIC ACID, Programmed cell death 1 ligand 2, SODIUM ION | | Authors: | Lazar-Molnar, E, Ramagopal, U, Cao, E, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-17 | | Release date: | 2008-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DP7
 
 | | CRYSTAL STRUCTURE OF SAM-dependent methyltransferase from Bacteroides vulgatus ATCC 8482 | | Descriptor: | SAM-dependent methyltransferase | | Authors: | Malashkevich, V.N, Toro, R, Ramagopal, U, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | CRYSTAL STRUCTURE OF SAM-dependent methyltransferase from Bacteroides vulgatus ATCC 8482
To be Published
|
|
3JU2
 
 | | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein SMc04130 | | Authors: | Patskovsky, Y, Foti, R, Ramagopal, U, Malashkevich, V, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF PROTEIN SMc04130 FROM Sinorhizobium meliloti
To be Published
|
|
3JVD
 
 | | Crystal structure of putative transcription regulation repressor (LACI family) FROM Corynebacterium glutamicum | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulators | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Foti, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-16 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of LACI family protein from Corynebacterium glutamicum
To be Published
|
|
3MOG
 
 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3MN1
 
 | | Crystal structure of probable yrbi family phosphatase from pseudomonas syringae pv.phaseolica 1448a | | Descriptor: | CHLORIDE ION, probable yrbi family phosphatase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3N28
 
 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3MY9
 
 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
2G64
 
 | | Structure of Caenorhabditis elegans 6-pyruvoyl tetrahydropterin synthase | | Descriptor: | GLYCEROL, Putative 6-pyruvoyl tetrahydrobiopterin synthase, SODIUM ION, ... | | Authors: | Wengerter, B.C, Patskovsky, Y, Zhan, C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans 6-Pyruvoyl Tetrahydropterin Synthase
To be Published
|
|
2FGS
 
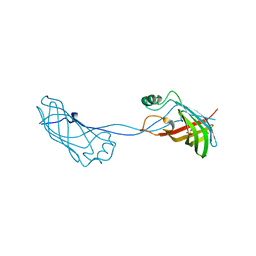 | | Crystal structure of Campylobacter jejuni YCEI protein, structural genomics | | Descriptor: | Putative periplasmic protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-22 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Campylobacter Jejuni YceI Periplasmic Protein
To be Published
|
|
2HC9
 
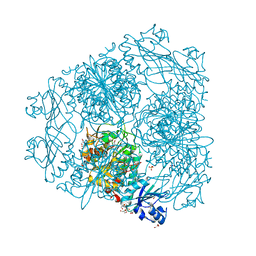 | | Structure of Caenorhabditis elegans leucine aminopeptidase-zinc complex (LAP1) | | Descriptor: | BICARBONATE ION, GLYCEROL, Leucine aminopeptidase 1, ... | | Authors: | Zhan, C, Patskovsky, Y, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Function of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
2HB6
 
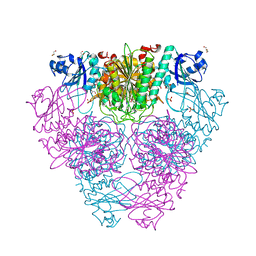 | | Structure of Caenorhabditis elegans leucine aminopeptidase (LAP1) | | Descriptor: | GLYCEROL, Leucine aminopeptidase 1, SODIUM ION, ... | | Authors: | Patskovsky, Y, Zhan, C, Wengerter, B.C, Ramagopal, U, Milstein, S, Vidal, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Leucine Aminopeptidase
To be Published
|
|
2HDW
 
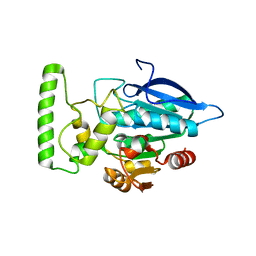 | | Crystal structure of hypothetical protein PA2218 from Pseudomonas Aeruginosa | | Descriptor: | Hypothetical protein PA2218 | | Authors: | Fedorov, A.A, Fedorov, E.V, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein PA2218
from Pseudomonas Aeruginosa
To be Published
|
|
2HF2
 
 | |
3HMU
 
 | | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi | | Descriptor: | Aminotransferase, class III, CHLORIDE ION, ... | | Authors: | Toro, R, Bonanno, J.B, Ramagopal, U, Freeman, J, Bain, K.T, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a class III aminotransferase from Silicibacter pomeroyi
To be Published
|
|
3GG9
 
 | | CRYSTAL STRUCTURE OF putative D-3-phosphoglycerate dehydrogenase oxidoreductase from Ralstonia solanacearum | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Putative D-3-Phosphoglycerate Dehydrogenase from Ralstonia Solanacearum
To be Published
|
|
3I4S
 
 | | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN blr8122 FROM Bradyrhizobium japonicum | | Descriptor: | GLYCEROL, HISTIDINE TRIAD PROTEIN | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN FROM Bradyrhizobium japonicum
To be Published
|
|
3FNR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | Descriptor: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-26 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
3HV2
 
 | | Crystal structure of signal receiver domain OF HD domain-containing protein FROM Pseudomonas fluorescens Pf-5 | | Descriptor: | Response regulator/HD domain protein, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of signal receiver domain oF HD domain-containing protein 3 FROM Pseudomonas fluorescens
To be Published
|
|
3FHL
 
 | | Crystal structure of a putative oxidoreductase from bacteroides fragilis nctc 9343 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative oxidoreductase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-09 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase from Bacteroides Fragilis Nctc 9343
To be Published
|
|
2ATM
 
 | | Crystal structure of the recombinant allergen Ves v 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hyaluronoglucosaminidase, SULFATE ION | | Authors: | Skov, L.K, Seppala, U, Coen, J.J.F, Crickmore, N, King, T.P, Monsalve, R, Kastrup, J.S, Spangfort, M.D, Gajhede, M. | | Deposit date: | 2005-08-25 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant Ves v 2 at 2.0 Angstrom resolution: structural analysis of an allergenic hyaluronidase from wasp venom.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1Y0G
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN, STRUCTURAL GENOMICS | | Descriptor: | 2-[(2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL]PHENOL, Protein yceI | | Authors: | Patskovsky, Y.V, Strokopytov, B, Ramagopal, U, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PERIPLASMIC PROTEIN
To be Published
|
|
