1G7D
 
 | | NMR STRUCTURE OF ERP29 C-DOMAIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, A, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1G7E
 
 | | NMR STRUCTURE OF N-DOMAIN OF ERP29 PROTEIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, M, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
2K7R
 
 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
1UCP
 
 | | NMR structure of the PYRIN domain of human ASC | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Liepinsh, E, Barbals, R, Dahl, E, Sharipo, A, Staub, E, Otting, G. | | Deposit date: | 2003-04-16 | | Release date: | 2003-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The death-domain fold of the ASC PYRIN domain, presenting a basis for PYRIN/PYRIN recognition
J.Mol.Biol., 332, 2003
|
|
1TUH
 
 | | Structure of Bal32a from a Soil-Derived Mobile Gene Cassette | | Descriptor: | ACETATE ION, hypothetical protein EGC068 | | Authors: | Robinson, A, Wu, P.S.-C, Harrop, S.J, Schaeffer, P.M, Dixon, N.E, Gillings, M.R, Holmes, A.J, Nevalainen, K.M.H, Otting, G, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Integron-associated Mobile Gene Cassettes Code for Folded Proteins: The Structure of Bal32a, a New Member of the Adaptable alpha+beta Barrel Family
J.Mol.Biol., 346, 2005
|
|
2D3J
 
 | |
2DDI
 
 | |
2AYA
 
 | |
1JBI
 
 | | NMR structure of the LCCL domain | | Descriptor: | cochlin | | Authors: | Liepinsh, E, Trexler, M, Kaikkonen, A, Weigelt, J, Banyai, L, Patthy, L, Otting, G. | | Deposit date: | 2001-06-05 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the LCCL domain and implications for DFNA9 deafness disorder.
EMBO J., 20, 2001
|
|
1J3G
 
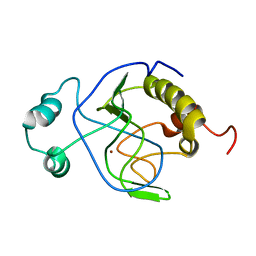 | | Solution structure of Citrobacter Freundii AmpD | | Descriptor: | AmpD protein, ZINC ION | | Authors: | Liepinsh, E, Genereux, C, Dehareng, D, Joris, B, Otting, G. | | Deposit date: | 2003-01-31 | | Release date: | 2003-02-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Citrobacter freundii AmpD, Comparison with Bacteriophage T7 Lysozyme
and Homology with PGRP Domains
J.Mol.Biol., 327, 2003
|
|
1J5B
 
 | | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein | | Descriptor: | Antifreeze protein type 1 analogue | | Authors: | Liepinsh, E, Otting, G, Harding, M.M, Ward, L.G, Mackay, J.P, Haymet, A.D. | | Deposit date: | 2002-03-22 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrophobic analogue of the winter flounder antifreeze protein.
Eur.J.Biochem., 269, 2002
|
|
1JWE
 
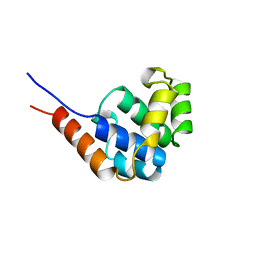 | | NMR Structure of the N-Terminal Domain of E. Coli Dnab Helicase | | Descriptor: | PROTEIN (DNAB HELICASE) | | Authors: | Weigelt, J, Brown, S.E, Miles, C.S, Dixon, N.E, Otting, G. | | Deposit date: | 1999-01-22 | | Release date: | 1999-01-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer.
Structure Fold.Des., 7, 1999
|
|
2DDJ
 
 | |
2XY8
 
 | |
1FTZ
 
 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
1SAN
 
 | |
7RNW
 
 | |
6WUP
 
 | |
6JPW
 
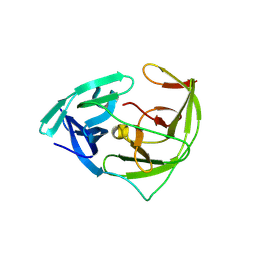 | | Crystal structure of Zika NS2B-NS3 protease with compound 1C | | Descriptor: | NS3 protease, SER-C0F-GLY-LYS-ARG-LYS, Serine protease subunit NS2B | | Authors: | Quek, J.P. | | Deposit date: | 2019-03-28 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Biocompatible Macrocyclization between Cysteine and 2-Cyanopyridine Generates Stable Peptide Inhibitors.
Org.Lett., 21, 2019
|
|
4MZ9
 
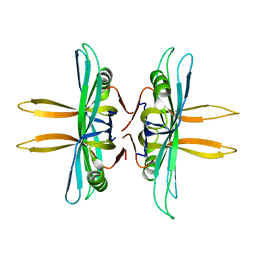 | | Revised structure of E. coli SSB | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Oakley, A.J. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intramolecular binding mode of the C-terminus of Escherichia coli single-stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy.
Nucleic Acids Res., 42, 2014
|
|
7R6O
 
 | | Pyrrolysyl-tRNA synthetase from methanogenic archaeon ISO4-G1 (G1PylRS) | | Descriptor: | 1,2-ETHANEDIOL, Pyrrolysyl-tRNA synthetase PylS, SULFATE ION | | Authors: | Frkic, R.L, Huber, T, Jackson, C.J. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic Encoding of Cyanopyridylalanine for In-Cell Protein Macrocyclization by the Nitrile-Aminothiol Click Reaction.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4GX9
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
4GX8
 
 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | CHLORIDE ION, DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Robinson, A, Horan, N, Xu, Z.-Q, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
2LFB
 
 | | HOMEODOMAIN FROM RAT LIVER LFB1/HNF1 TRANSCRIPTION FACTOR, NMR, 20 STRUCTURES | | Descriptor: | LFB1/HNF1 TRANSCRIPTION FACTOR | | Authors: | Schott, O, Billeter, M, Leiting, B, Wider, G, Wuthrich, K. | | Deposit date: | 1996-12-12 | | Release date: | 1997-03-12 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the non-classical homeodomain from the rat liver LFB1/HNF1 transcription factor.
J.Mol.Biol., 267, 1997
|
|
