1NGR
 
 | |
1NKL
 
 | |
1F4K
 
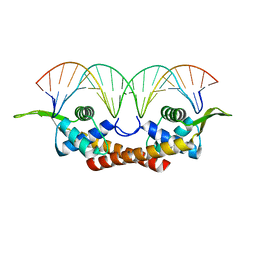 | | CRYSTAL STRUCTURE OF THE REPLICATION TERMINATOR PROTEIN/B-SITE DNA COMPLEX | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*AP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*GP*AP*AP*CP*AP*TP*TP*AP*TP*GP*TP*TP*CP*AP*TP*AP*G)-3', REPLICATION TERMINATION PROTEIN | | Authors: | Wilce, J.A, Vivian, J.P, Hastings, A.F, Otting, G, Folmer, R.H.A, Duggin, I.G, Wake, R.G, Wilce, M.C.J. | | Deposit date: | 2000-06-08 | | Release date: | 2001-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest
Nat.Struct.Biol., 8, 2001
|
|
1ZFO
 
 | | AMINO-TERMINAL LIM-DOMAIN PEPTIDE OF LASP-1, NMR | | Descriptor: | LASP-1, ZINC ION | | Authors: | Hammarstrom, A, Berndt, K.D, Sillard, R, Adermann, K, Otting, G. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit.
Biochemistry, 35, 1996
|
|
1AHD
 
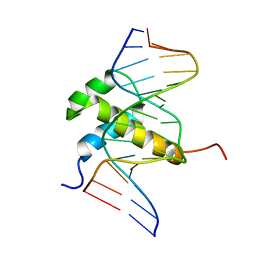 | | DETERMINATION OF THE NMR SOLUTION STRUCTURE OF AN ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), DNA (5'-D(*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), Homeotic protein antennapedia | | Authors: | Billeter, M, Qian, Y.Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex.
J.Mol.Biol., 234, 1993
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
2HOA
 
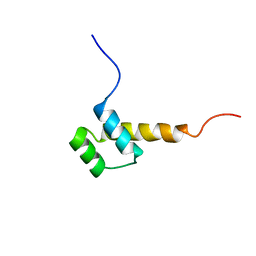 | | STRUCTURE DETERMINATION OF THE ANTP(C39->S) HOMEODOMAIN FROM NUCLEAR MAGNETIC RESONANCE DATA IN SOLUTION USING A NOVEL STRATEGY FOR THE STRUCTURE CALCULATION WITH THE PROGRAMS DIANA, CALIBA, HABAS AND GLOMSA | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Guntert, P, Qian, Y.-Q, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1992-04-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure determination of the Antp (C39----S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA.
J.Mol.Biol., 217, 1991
|
|
1FOV
 
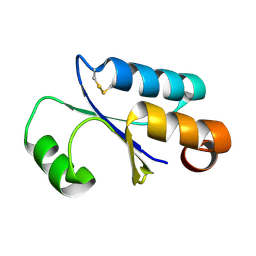 | | GLUTAREDOXIN 3 FROM ESCHERICHIA COLI IN THE FULLY OXIDIZED FORM | | Descriptor: | GLUTAREDOXIN 3 | | Authors: | Nordstrand, K, Sandstrom, A, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 2000-08-29 | | Release date: | 2000-10-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of oxidized glutaredoxin 3 from Escherichia coli.
J.Mol.Biol., 303, 2000
|
|
7N3J
 
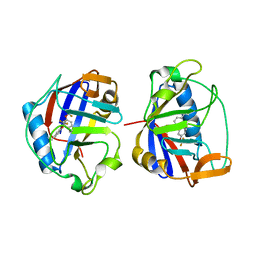 | |
6O6I
 
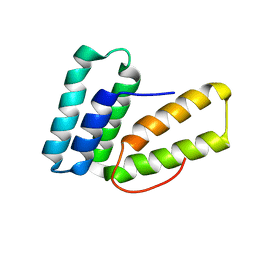 | |
1HOM
 
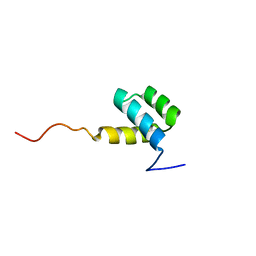 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
1T3W
 
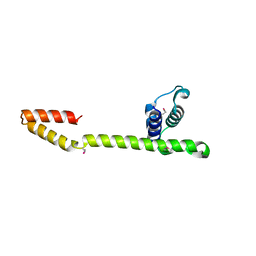 | | Crystal Structure of the E.coli DnaG C-terminal domain (residues 434 to 581) | | Descriptor: | ACETIC ACID, DNA primase | | Authors: | Oakley, A.J, Loscha, K.V, Schaeffer, P.M, Liepinsh, E, Wilce, M.C.J, Otting, G, Dixon, N.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal and solution structures of the helicase-binding domain of Escherichia coli primase
J.Biol.Chem., 280, 2005
|
|
1G7D
 
 | | NMR STRUCTURE OF ERP29 C-DOMAIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, A, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
1G7E
 
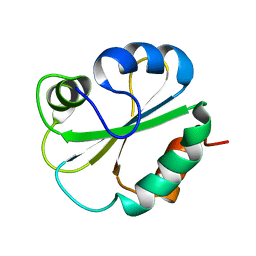 | | NMR STRUCTURE OF N-DOMAIN OF ERP29 PROTEIN | | Descriptor: | ENDOPLASMIC RETICULUM PROTEIN ERP29 | | Authors: | Liepinsh, E, Mkrtchian, S, Barishev, M, Sharipo, M, Ingelman-Sundberg, M, Otting, G. | | Deposit date: | 2000-11-10 | | Release date: | 2000-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Thioredoxin fold as homodimerization module in the putative chaperone ERp29: NMR structures of the domains and experimental model of the 51 kDa dimer.
Structure, 9, 2001
|
|
7RFD
 
 | |
2LRR
 
 | | Solution structure of the R3H domain from human Smubp-2 in complex with 2'-deoxyguanosine-5'-monophosphate | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-binding protein SMUBP-2 | | Authors: | Jaudzems, K, Zhulenkovs, D, Otting, G, Liepinsh, E. | | Deposit date: | 2012-04-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for 5'-End-Specific Recognition of Single-Stranded DNA by the R3H Domain from Human Smubp-2
J.Mol.Biol., 12, 2012
|
|
1ADR
 
 | |
1AOY
 
 | |
1AXJ
 
 | |
1MSZ
 
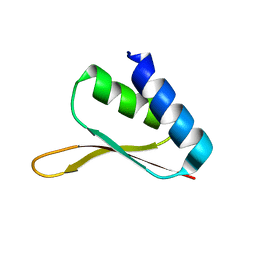 | | Solution structure of the R3H domain from human Smubp-2 | | Descriptor: | DNA-binding protein SMUBP-2 | | Authors: | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
1PCE
 
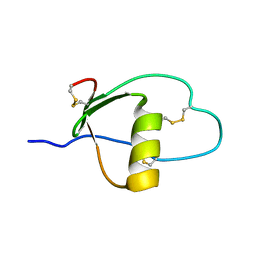 | | SOLUTION STRUCTURE AND DYNAMICS OF PEC-60, A PROTEIN OF THE KAZAL TYPE INHIBITOR FAMILY, DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | PEC-60 | | Authors: | Liepinsh, E, Berndt, K.D, Sillard, R, Mutt, V, Otting, G. | | Deposit date: | 1994-02-22 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of PEC-60, a protein of the Kazal type inhibitor family, determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 239, 1994
|
|
2M66
 
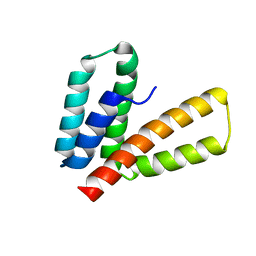 | | Endoplasmic reticulum protein 29 (ERp29) C-terminal domain: 3D Protein Fold Determination from Backbone Amide Pseudocontact Shifts Generated by Lanthanide Tags at Multiple Sites | | Descriptor: | Endoplasmic reticulum resident protein 29 | | Authors: | Yagi, H, Pilla, K, Maleckis, A, Graham, B, Huber, T, Otting, G. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites
Structure, 21, 2013
|
|
1UAP
 
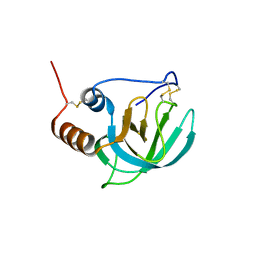 | | NMR structure of the NTR domain from human PCOLCE1 | | Descriptor: | Procollagen C-proteinase enhancer protein | | Authors: | Liepinsh, E, Banyai, L, Pintacuda, G, Trexler, M, Patthy, L, Otting, G. | | Deposit date: | 2003-03-14 | | Release date: | 2003-07-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Netrin-like Domain (NTR) of Human Type I Procollagen C-Proteinase Enhancer Defines Structural Consensus of NTR Domains and Assesses Potential Proteinase Inhibitory Activity and Ligand Binding.
J.Biol.Chem., 278, 2003
|
|
2HAJ
 
 | |
2K7R
 
 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
