4D6O
 
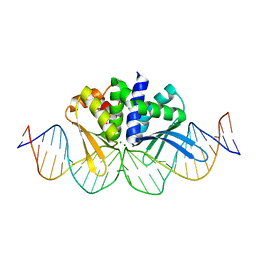 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 1H INCUBATION IN 5MM MG (STATE 2) | | Descriptor: | 25MER, ACETATE ION, HOMING ENDONUCLEASE I-DMOI, ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4D6N
 
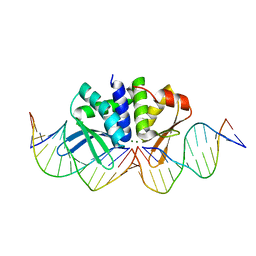 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 10 DAYS INCUBATION IN 5MM MG (STATE 7) | | Descriptor: | 5'-D(*DCP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*DCP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*DAP*C)-3', 5'-D(*DGP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*DAP)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-11-13 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2KP4
 
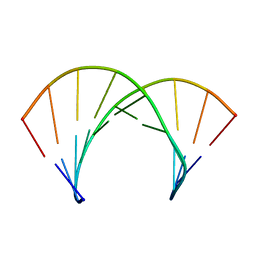 | | Structure of 2'F-ANA/RNA hybrid duplex | | Descriptor: | DNA (5'-D(*(GFL)P*(CFL)P*(TAF)P*(A5L)P*(TAF)P*(A5L)P*(A5L)P*(TAF)P*(GFL)P*(GFL))-3'), RNA (5'-R(*CP*CP*AP*UP*UP*AP*UP*AP*GP*C)-3') | | Authors: | Gonzalez, C, Martin-Pintado, N, Watts, J, Gomez-Pinto, I, Dhama, M, Orozco, M, Schwartzentruber, J, Portella, G. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Differential stability of 2'F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility.
Nucleic Acids Res., 38, 2010
|
|
2KP3
 
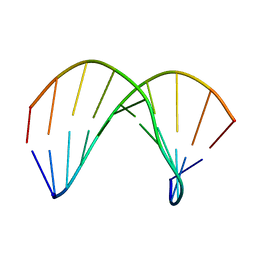 | | Structure of ANA-RNA hybrid duplex | | Descriptor: | RNA (5'-R(*(GAO)P*(CAR)P*(UAR)P*(A5O)P*(UAR)P*(A5O)P*(A5O)P*(UAR)P*(GAO)P*(GAO))-3'), RNA (5'-R(*CP*CP*AP*UP*UP*AP*UP*AP*GP*C)-3') | | Authors: | Gonzalez, C, Martn-Pintado, N, Watts, J, Gomez-Pinto, I, Dhama, M, Orozco, M, Schwartzentruber, J, Portella, G. | | Deposit date: | 2009-10-06 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Differential stability of 2'F-ANA*RNA and ANA*RNA hybrid duplexes: roles of structure, pseudohydrogen bonding, hydration, ion uptake and flexibility.
Nucleic Acids Res., 38, 2010
|
|
5M33
 
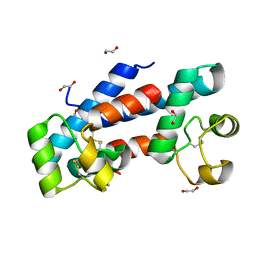 | | Structural tuning of CD81LEL (space group P21) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M2C
 
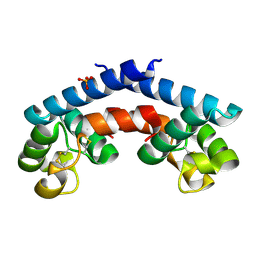 | | Structural tuning of CD81LEL (space group P32 1 2) | | Descriptor: | CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
2M8A
 
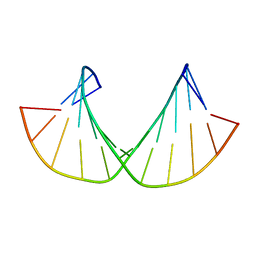 | | 2'F-ANA/2'F-RNA alternated sequences | | Descriptor: | 2'F-RNA/2'F-ANA chimeric duplex | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2M84
 
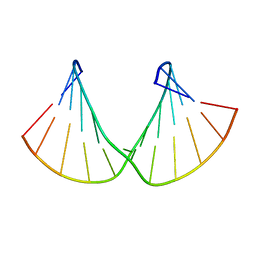 | | Structure of 2'F-RNA/2'F-ANA chimeric duplex | | Descriptor: | 2'F-RNA/2'F-ANA CHIMERIC DUPLEX | | Authors: | Martin-Pintado, N, Deleavey, G, Portella, G, Campos, R, Orozco, M, Damha, M, Gonzalez, C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone FCHO Hydrogen Bonds in 2'F-Substituted Nucleic Acids.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
8BQY
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
3OB6
 
 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | Descriptor: | ARGININE, AdiC arginine:agmatine antiporter | | Authors: | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1E66
 
 | | STRUCTURE OF ACETYLCHOLINESTERASE COMPLEXED WITH (-)-HUPRINE X AT 2.1A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CHLORO-9-ETHYL-6,7,8,9,10,11-HEXAHYDRO-7,11-METHANOCYCLOOCTA[B]QUINOLIN-12-AMINE, ACETYLCHOLINESTERASE | | Authors: | Dvir, H, Harel, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-08-08 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 3D Structure of Torpedo Californica Acetylcholinesterase Complexed with Huprine X at 2. 1 A Resolution: Kinetic and Molecular Dynamic Correlates.
Biochemistry, 41, 2002
|
|
6HB4
 
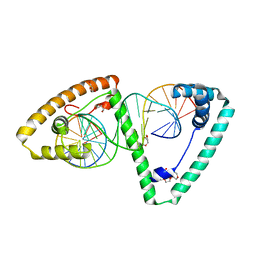 | | TFAM in Complex with Site-Y | | Descriptor: | DNA (5'*CP*TP*GP*TP*GP*CP*AP*GP*AP*CP*AP*TP*TP*CP*AP*AP*TP*TP*GP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*CP*AP*AP*TP*TP*GP*AP*AP*TP*GP*TP*CP*TP*GP*CP*AP*CP*AP*G)-3'), GLYCEROL, ... | | Authors: | Cuppari, A, Fernandez-Millan, P, Rubio-Cosials, A, Tarres-Sole, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
6HC3
 
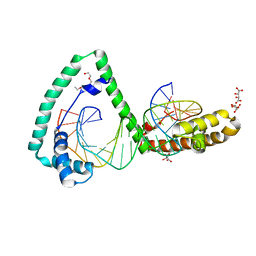 | | TFAM bound to Site-X | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*GP*TP*GP*GP*AP*AP*AP*TP*TP*TP*TP*TP*TP*GP*TP*TP*AP*G)-3'), DNA/RNA (5'-D(*TP*AP*AP*CP*AP*AP*AP*AP*AP*AP*TP*TP*TP*CP*CP*AP*CP*CP*AP*AP*AP*C)-3'), L(+)-TARTARIC ACID, ... | | Authors: | Fernandez-Millan, P, Cuppari, A, Tarres-Sole, A, Rubio-Cosials, A, Lyonnais, S, Sola, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DNA specificities modulate the binding of human transcription factor A to mitochondrial DNA control region.
Nucleic Acids Res., 47, 2019
|
|
5JGH
 
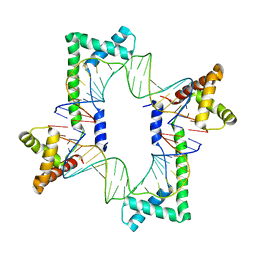 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.6 Angstrom resolution | | Descriptor: | ACETATE ION, ARS-binding factor 2, mitochondrial, ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
5JH0
 
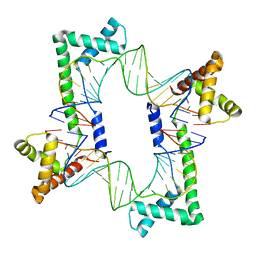 | | Crystal structure of the mitochondrial DNA packaging protein Abf2p in complex with DNA at 2.18 Angstrom resolution | | Descriptor: | ARS-binding factor 2, mitochondrial, DNA (5'-D(*AP*AP*TP*AP*AP*TP*AP*AP*AP*TP*TP*AP*TP*AP*TP*AP*AP*TP*AP*TP*AP*A)-3'), ... | | Authors: | Chakraborty, A, Lyonnais, S, Sola, M. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Nucleic Acids Res., 45, 2017
|
|
2DH3
 
 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ZINC ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
2DH2
 
 | | Crystal Structure of human ED-4F2hc | | Descriptor: | 4F2 cell-surface antigen heavy chain, ACETATE ION | | Authors: | Fort, J, Fita, I, Palacin, M. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane.
J.Biol.Chem., 282, 2007
|
|
4U87
 
 | | Crystal structure of the Ba-soaked C2 crystal form of pMV158 replication initiator RepB (P3221 space group) | | Descriptor: | BARIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Boer, D.R, Ruiz Maso, J.A, del Solar, G, Coll, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Conformational plasticity of RepB, the replication initiator protein of promiscuous streptococcal plasmid pMV158.
Sci Rep, 6, 2016
|
|
5N2Q
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to 26nt pMV158 oriT DNA | | Descriptor: | CHLORIDE ION, DNA (26-MER), GLYCEROL, ... | | Authors: | Russi, S, Boer, D.R, Coll, M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7PVU
 
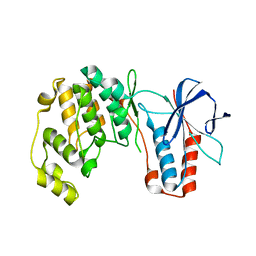 | | Crystal structure of p38alpha C162S in complex with CAS2094511-69-8, P 1 21 1 | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-cyclobutyl-1H-1,3-benzodiazol-5-yl)-2-fluorobenzene-1-sulfonamide | | Authors: | Baginski, B, Pous, J, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2021-10-05 | | Release date: | 2023-05-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Characterization of p38 alpha autophosphorylation inhibitors that target the non-canonical activation pathway.
Nat Commun, 14, 2023
|
|
4LVK
 
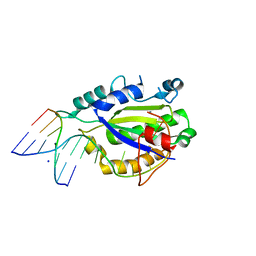 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt+3'Phosphate). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGpo oligonucleotide, MANGANESE (II) ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVM
 
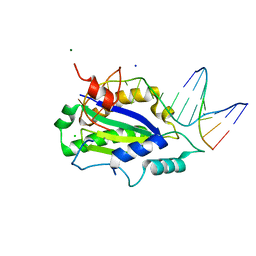 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (23nt). Mn-bound crystal structure at pH 6.5 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTGT oligonucleotide, CHLORIDE ION, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVJ
 
 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 5.5 | | Descriptor: | ACETATE ION, ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LVI
 
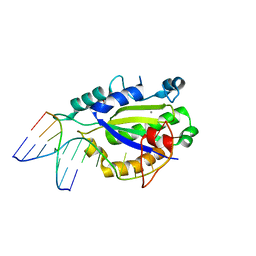 | | MobM Relaxase Domain (MOBV; Mob_Pre) bound to plasmid pMV158 oriT DNA (22nt). Mn-bound crystal structure at pH 4.6 | | Descriptor: | ACTTTAT oligonucleotide, ATAAAGTATAGTGTG oligonucleotide, GLYCEROL, ... | | Authors: | Pluta, R, Boer, D.R, Coll, M. | | Deposit date: | 2013-07-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of a histidine-DNA nicking/joining mechanism for gene transfer and promiscuous spread of antibiotic resistance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
