2CYY
 
 | | Crystal structure of PH1519 from Pyrococcus horikosii OT3 | | Descriptor: | CALCIUM ION, GLUTAMINE, Putative HTH-type transcriptional regulator PH1519 | | Authors: | Okazaki, N, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-08 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PH1519 from Pyrococcus horikosii OT3
To be Published
|
|
2DSY
 
 | | Crystal structure of TTHA0281 from thermus thermophilus HB8 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical protein TTHA0281, MAGNESIUM ION | | Authors: | Okazaki, N, Kumei, M, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-07 | | Release date: | 2007-03-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a UPF0150-family protein from Thermus thermophilus HB8
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
2EA9
 
 | |
2EJ5
 
 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
3VGH
 
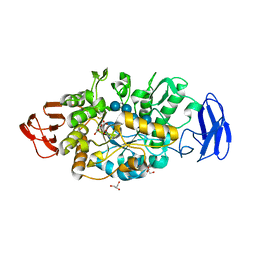 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGG
 
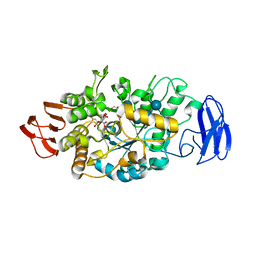 | | Crystal structure of glycosyltrehalose trehalohydrolase (E283Q) complexed with maltoheptaose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGB
 
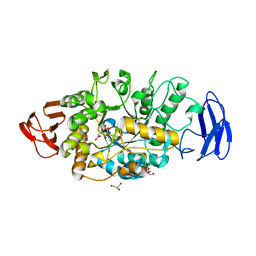 | | Crystal structure of glycosyltrehalose trehalohydrolase (GTHase) from Sulfolobus solfataricus KM1 | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGE
 
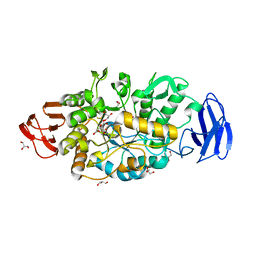 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGD
 
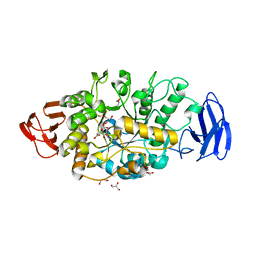 | | Ctystal structure of glycosyltrehalose trehalohydrolase (D252E) | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
3VGF
 
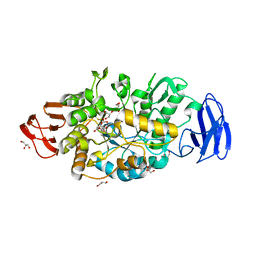 | | Crystal structure of glycosyltrehalose trehalohydrolase (D252S) complexed with maltotriosyltrehalose | | Descriptor: | CITRATE ANION, GLYCEROL, Malto-oligosyltrehalose trehalohydrolase, ... | | Authors: | Okazaki, N, Tamada, T, Feese, M.D, Kato, M, Miura, Y, Komeda, T, Kobayashi, K, Kondo, K, Kuroki, R. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition mechanism of a glycosyltrehalose trehalohydrolase from Sulfolobus solfataricus KM1.
Protein Sci., 21, 2012
|
|
5ZCR
 
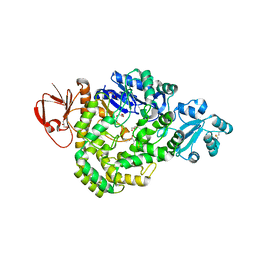 | | DSM5389 glycosyltrehalose synthase | | Descriptor: | GLYCEROL, MAGNESIUM ION, Maltooligosyl trehalose synthase | | Authors: | Tamada, T, Okazaki, N. | | Deposit date: | 2018-02-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of glycosyltrehalose synthase from Sulfolobus shibatae DSM5389
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2DJW
 
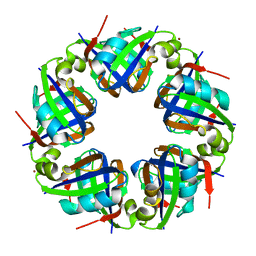 | | Crystal structure of TTHA0845 from Thermus thermophilus HB8 | | Descriptor: | ZINC ION, probable transcriptional regulator, AsnC family | | Authors: | Okazaki, N, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the stand-alone RAM-domain protein from Thermus thermophilus HB8
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
3VGS
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGT
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGU
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
3VGV
 
 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
1V40
 
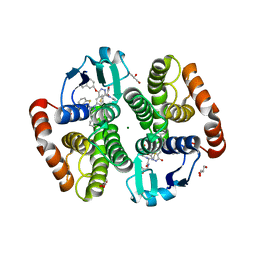 | | First Inhibitor Complex Structure of Human Hematopoietic Prostaglandin D Synthase | | Descriptor: | 3-(1,3-BENZOTHIAZOL-2-YL)-2-(1,4-DIOXO-1,2,3,4-TETRAHYDROPHTHALAZIN-6-YL)-5-[(E)-2-PHENYLVINYL]-3H-TETRAAZOL-2-IUM, GLUTATHIONE, GLYCEROL, ... | | Authors: | Inoue, T, Okano, Y, Kado, Y, Aritake, K, Irikura, D, Uodome, N, Kinugasa, S, Okazaki, N, Matsumura, H, Kai, Y, Urade, Y. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First determination of the inhibitor complex structure of human hematopoietic prostaglandin D synthase.
J.Biochem.(Tokyo), 135, 2004
|
|
3FX5
 
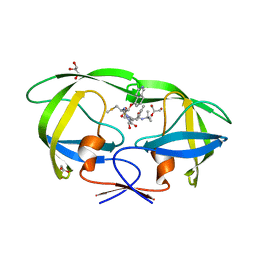 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
2EO8
 
 | |
3W6E
 
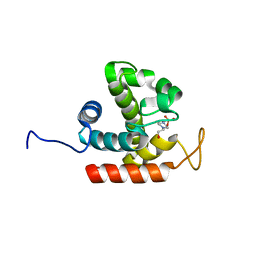 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6D
 
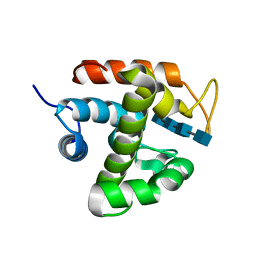 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6B
 
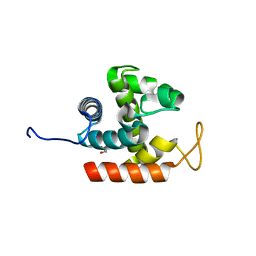 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 | | Descriptor: | GLYCEROL, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6F
 
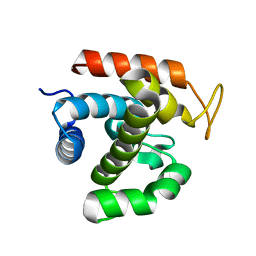 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E162Q) in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
3W6C
 
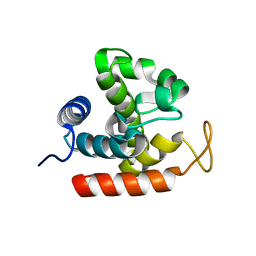 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 in complex with disaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
2ZYE
 
 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by Neutron Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | NEUTRON DIFFRACTION (1.9 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
