2K6Q
 
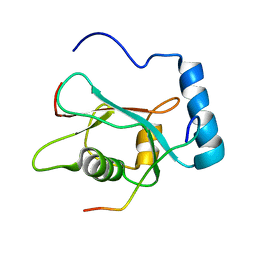 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
4DVZ
 
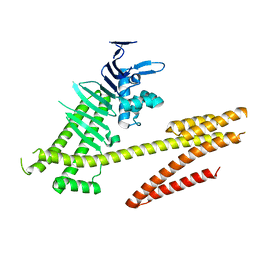 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
4DVY
 
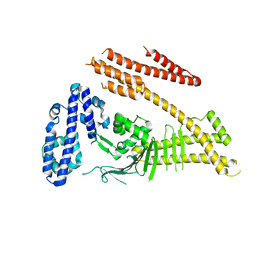 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
2KZK
 
 | | Solution structure of alpha-mannosidase binding domain of Atg34 | | Descriptor: | Uncharacterized protein YOL083W | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-18 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2KZB
 
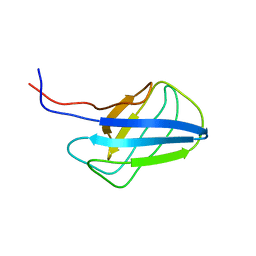 | | Solution structure of alpha-mannosidase binding domain of Atg19 | | Descriptor: | Autophagy-related protein 19 | | Authors: | Watanabe, Y, Noda, N, Kumeta, H, Suzuki, K, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34.
J.Biol.Chem., 285, 2010
|
|
2D5R
 
 | | Crystal Structure of a Tob-hCaf1 Complex | | Descriptor: | CCR4-NOT transcription complex subunit 7, Tob1 protein | | Authors: | Horiuchi, M, Suzuki, N.N, Muroya, N, Takahasi, K, Nishida, M, Yoshida, Y, Ikematsu, N, Nakamura, T, Kawamura-Tsuzuku, J, Yamamoto, T, Inagaki, F. | | Deposit date: | 2005-11-04 | | Release date: | 2006-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the antiproliferative activity of the Tob-hCaf1 complex.
J.Biol.Chem., 284, 2009
|
|
4WHM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REN
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | Descriptor: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | Descriptor: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2014-09-23 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
1J2F
 
 | | X-ray crystal structure of IRF-3 and its functional implications | | Descriptor: | Interferon regulatory factor 3 | | Authors: | Takahasi, K, Noda, N, Horiuchi, M, Mori, M, Okabe, Y, Fukuhara, Y, Terasawa, H, Fujita, T, Inagaki, F. | | Deposit date: | 2003-01-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of IRF-3 and its functional implications.
Nat.Struct.Biol., 10, 2003
|
|
3WC4
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | Descriptor: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | Authors: | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
6KXM
 
 | |
