1EWT
 
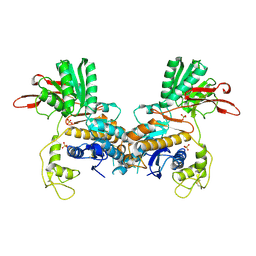 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1, SULFATE ION | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1ARP
 
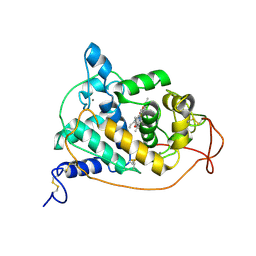 | |
3VNH
 
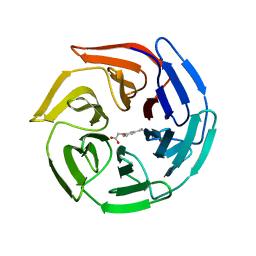 | | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 Soaked with Synthetic Small Molecular
To be Published
|
|
1EWV
 
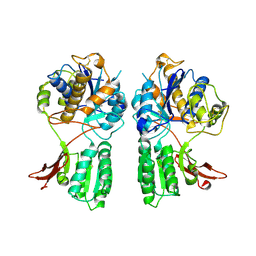 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 LIGAND FREE FORM II | | Descriptor: | METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 | | Authors: | Kunishima, N, Shimada, Y, Tsuji, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-27 | | Release date: | 2000-12-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
1EWK
 
 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | Authors: | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | Deposit date: | 2000-04-26 | | Release date: | 2000-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
3VNG
 
 | | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization | | Descriptor: | 2-(3-((3-(5-(furan-2-yl)-1,3,4-oxadiazol-2-yl)ureido)methyl)phenoxy)acetic acid, Kelch-like ECH-associated protein 1 | | Authors: | Kunishima, N, Tanaka, T, Satoh, M, Saburi, H. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Keap1 in Complex with Synthetic Small Molecular based on a co-crystallization
To be Published
|
|
1J33
 
 | |
1J1Y
 
 | | Crystal Structure of PaaI from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PaaI protein | | Authors: | Kunishima, N, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-24 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1UAY
 
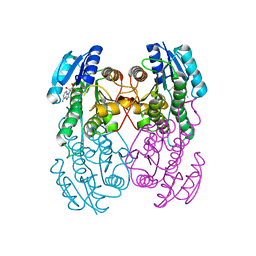 | | Crystal Structure of Type II 3-Hydroxyacyl-CoA Dehydrogenase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE, Type II 3-hydroxyacyl-CoA dehydrogenase | | Authors: | Kunishima, N, Asada, Y, Yokoyama, S, Kuramitsu, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-03-25 | | Release date: | 2003-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Type II 3-Hydroxyacyl-CoA Dehydrogenase from Thermus thermophilus HB8
To be Published
|
|
1V8H
 
 | |
1VCE
 
 | |
1WLV
 
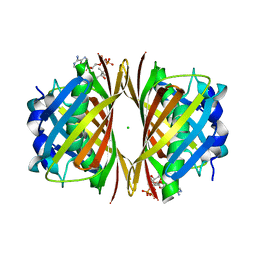 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1WLU
 
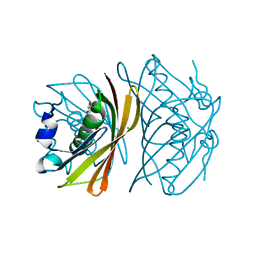 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, phenylacetic acid degradation protein PaaI | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1WN3
 
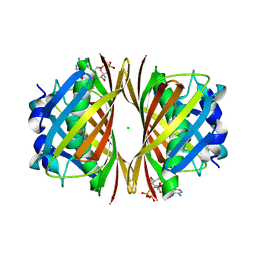 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, HEXANOYL-COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
6KJO
 
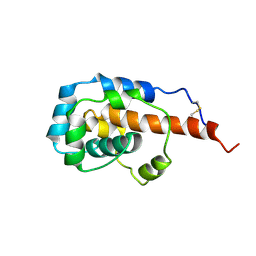 | | The microtubule-binding domains of yeast cytoplasmic dynein in the low affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6KJN
 
 | | The microtubule-binding domains of yeast cytoplasmic dynein in the high affinity state | | Descriptor: | Dynein heavy chain, cytoplasmic | | Authors: | Nishida, N, Komori, Y, Takarada, O, Watanabe, A, Tamura, S, Kubo, S, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
5WR2
 
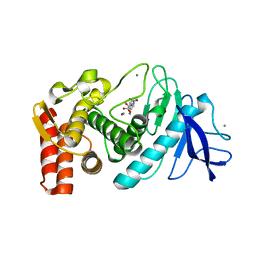 | | Thermolysin, SFX liganded form with oil-based carrier | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, Thermolysin, ... | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1GZB
 
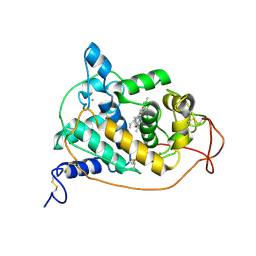 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
5WR3
 
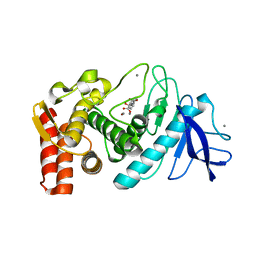 | | Thermolysin, SFX liganded form with water-based carrier | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, Thermolysin, ... | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WR6
 
 | | Thermolysin, liganded form with cryo condition 2 | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, Thermolysin, ... | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ZGO
 
 | |
5WR4
 
 | | Thermolysin, SFX unliganded form with oil-based carrier | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5WR5
 
 | | Thermolysin, liganded form with cryo condition 1 | | Descriptor: | CALCIUM ION, N-[(benzyloxy)carbonyl]-L-aspartic acid, TETRAETHYLENE GLYCOL, ... | | Authors: | Kunishima, N, Naitow, H, Matsuura, Y. | | Deposit date: | 2016-11-29 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-ligand complex structure from serial femtosecond crystallography using soaked thermolysin microcrystals and comparison with structures from synchrotron radiation
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1WNG
 
 | |
1WM6
 
 | |
