5KR4
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR6
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
1QP8
 
 | |
5KQU
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQT
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminoburyrate transaminase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR3
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KQW
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 1,2-ETHANEDIOL, 4-aminobutyrate transaminase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-06 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
5KR5
 
 | | Directed Evolution of Transaminases By Ancestral Reconstruction. Using Old Proteins for New Chemistries | | 分子名称: | 4-aminobutyrate transaminase, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wilding, M, Newman, J, Peat, T.S, Scott, C. | | 登録日 | 2016-07-07 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Reverse engineering: transaminase biocatalyst development using ancestral sequence reconstruction
Green Chemistry, 19, 2017
|
|
6D63
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | 分子名称: | 3-oxopentanedioic acid, atzH | | 著者 | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | 登録日 | 2018-04-19 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
6HRH
 
 | | Structure of human erythroid-specific 5'-aminolevulinate synthase, ALAS2 | | 分子名称: | 5-aminolevulinate synthase, erythroid-specific, mitochondrial, ... | | 著者 | Bailey, H.J, Shrestha, L, Rembeza, E, Newman, J, Kupinska, K, Diaz-saez, L, Kennedy, E, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | 登録日 | 2018-09-27 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of human erythroid-specific 5'-aminolevulinate synthase, ALAS2
To Be Published
|
|
6I80
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT53 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 2 | | 著者 | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | Crystal Structure of the second bromodomain of BRD2 in complex with RT53
To Be Published
|
|
6I7X
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT53 | | 分子名称: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 4 | | 著者 | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Crystal Structure of the first bromodomain of BRD4 in complex with RT53
To Be Published
|
|
6I81
 
 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT56 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | 著者 | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal Structure of the second bromodomain of BRD2 in complex with RT56
To Be Published
|
|
6I7Y
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT56 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [1-[4-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]phenyl]piperidin-4-yl]-trimethyl-azanium | | 著者 | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Crystal Structure of the first bromodomain of BRD4 in complex with RT56
To Be Published
|
|
6PII
 
 | | The evolving story of AtzT, a periplasmic binding protein | | 分子名称: | Atrazine periplasmic binding protein, CALCIUM ION, ETHYL MERCURY ION, ... | | 著者 | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | 登録日 | 2019-06-26 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PI5
 
 | | The evolving story of AtzT, a periplasmic binding protein | | 分子名称: | Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE, GUANINE | | 著者 | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | 登録日 | 2019-06-26 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PI6
 
 | | The evolving story of AtzT, a periplasmic binding protein | | 分子名称: | 4-(ethylamino)-6-[(propan-2-yl)amino]-1,3,5-triazin-2-ol, Atrazine periplasmic binding protein, DIMETHYL SULFOXIDE | | 著者 | Peat, T.S, Newman, J, Scott, C, Esquirol, L, Dennis, M, Nebl, T. | | 登録日 | 2019-06-26 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The evolving story of AtzT, a periplasmic binding protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6R7T
 
 | | Crystal Structure of human Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody | | 分子名称: | Melanoma-associated antigen B1, anti MAGEB1 nanobody | | 著者 | Ye, M, Newman, J, Pike, A.C.W, Burgess-Brown, N, Cooper, C.D.O, Bountra, C, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F. | | 登録日 | 2019-03-29 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.682 Å) | | 主引用文献 | Crystal Structure of Melanoma-associated antigen B1 (MAGEB1) in complex with nanobody
To Be Published
|
|
3NF7
 
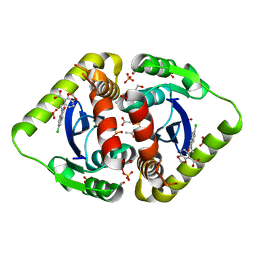 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF8
 
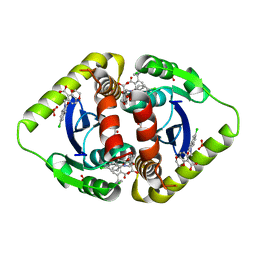 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 6-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NFA
 
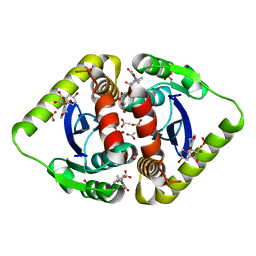 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 6-[(5-bromo-2,3-dioxo-2,3-dihydro-1H-indol-1-yl)methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETIC ACID, Integrase, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-10 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF6
 
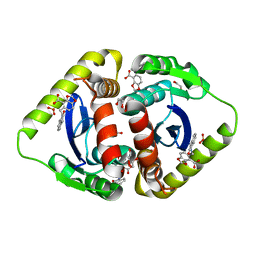 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NF9
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | 分子名称: | 1,2-ETHANEDIOL, 5-[(6-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | 登録日 | 2010-06-10 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
5G26
 
 | | Unveiling the Mechanism Behind the in-meso Crystallization of Membrane Proteins | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, INTIMIN | | 著者 | Zabara, A, Newman, J, Peat, T.S. | | 登録日 | 2016-04-07 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | The Nanoscience Behind the Art of in-Meso Crystallization of Membrane Proteins.
Nanoscale, 9, 2017
|
|
5HY4
 
 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | 分子名称: | Ring-opening amidohydrolase | | 著者 | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | 登録日 | 2016-02-01 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
