3VN0
 
 | |
3VRR
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) in complex with phospho-EGFR peptide | | Descriptor: | CALCIUM ION, Epidermal growth factor receptor, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VQG
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | C-terminal peptide from Immunoglobulin superfamily member 5, E3 ubiquitin-protein ligase LNX, SULFATE ION | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
3VRQ
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c (PL mutant) | | Descriptor: | CALCIUM ION, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VRO
 
 | | Crystal structure of the tyrosine kinase binding domain of Cbl-c in complex with phospho-Src peptide | | Descriptor: | CALCIUM ION, Proto-oncogene tyrosine-protein kinase Src, Signal transduction protein CBL-C | | Authors: | Takeshita, K, Tezuka, T, Isozaki, Y, Yamashita, E, Suzuki, M, Yamanashi, Y, Yamamoto, T, Nakagawa, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural flexibility regulates phosphopeptide-binding activity of the tyrosine kinase binding domain of Cbl-c.
J.Biochem., 152, 2012
|
|
3VTN
 
 | | The crystal structure of the C-terminal domain of Mu phage central spike - Pt derivative for MAD | | Descriptor: | FE (III) ION, PLATINUM (II) ION, Protein gp45 | | Authors: | Harada, K, Yamashita, E, Nakagawa, A, Takeda, S. | | Deposit date: | 2012-06-01 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the C-terminal domain of Mu phage central spike and functions of bound calcium ion
Biochim.Biophys.Acta, 1834, 2013
|
|
3VYI
 
 | |
3AXY
 
 | | Structure of Florigen Activation Complex Consisting of Rice Florigen Hd3a, 14-3-3 Protein GF14 and Rice FD Homolog OsFD1 | | Descriptor: | 14-3-3-like protein GF14-C, Protein HEADING DATE 3A, Rice FD homolog OsFD1 | | Authors: | Ohki, I, Furuita, K, Hayashi, K, Taoka, K, Tsuji, H, Nakagawa, A, Shimamoto, K, Kojima, C. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen
Nature, 476, 2011
|
|
1UHI
 
 | | Crystal structure of i-aequorin | | Descriptor: | (2R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(4-IODOBENZYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHH
 
 | | Crystal structure of cp-aequorin | | Descriptor: | (8R)-8-(CYCLOPENTYLMETHYL)-2-HYDROPEROXY-2-(4-HYDROXYBENZYL)-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H) -ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHJ
 
 | | Crystal structure of br-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-(4-BROMOBENZYL)-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1UHK
 
 | | Crystal structure of n-aequorin | | Descriptor: | (2S,8R)-8-BENZYL-2-HYDROPEROXY-6-(4-HYDROXYPHENYL)-2-(2-NAPHTHYLMETHYL)-7,8-DIHYDROIMIDAZO[1,2-A]PYRAZIN-3(2H)-ONE, Aequorin 2 | | Authors: | Toma, S, Chong, K.T, Nakagawa, A, Teranishi, K, Inouye, S, Shimomura, O. | | Deposit date: | 2003-07-03 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structures of semi-synthetic aequorins
Protein Sci., 14, 2005
|
|
1VF7
 
 | | Crystal structure of the membrane fusion protein, MexA of the multidrug transporter | | Descriptor: | Multidrug resistance protein mexA | | Authors: | Akama, H, Matsuura, T, Kashiwagi, S, Yoneyama, H, Tsukihara, T, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-04-09 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
3AWE
 
 | | Crystal structure of Pten-like domain of Ci-VSP (248-576) | | Descriptor: | ACETIC ACID, SODIUM ION, SULFATE ION, ... | | Authors: | Matsuda, M, Sakata, S, Takeshita, K, Suzuki, M, Yamashita, E, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-03-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity
J.Biol.Chem., 286, 2011
|
|
3AV4
 
 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AQJ
 
 | | Crystal Structure of a C-terminal domain of the bacteriophage P2 tail spike protein, gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Takeda, S, Yamashita, E, Nakagawa, A. | | Deposit date: | 2010-11-06 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | The host-binding domain of the P2 phage tail spike reveals a trimeric iron-binding structure
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3AV5
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AWF
 
 | | Crystal structure of Pten-like domain of Ci-VSP (236-576) | | Descriptor: | GLYCEROL, SULFATE ION, Voltage-sensor containing phosphatase | | Authors: | Matsuda, M, Sakata, S, Takeshita, K, Suzuki, M, Yamashita, E, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-03-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity
J.Biol.Chem., 286, 2011
|
|
3AWG
 
 | | Crystal structure of Pten-like domain of Ci-VSP G356A mutant (248-576) | | Descriptor: | SULFATE ION, Voltage-sensor containing phosphatase | | Authors: | Matsuda, M, Sakata, S, Takeshita, K, Suzuki, M, Yamashita, E, Okamura, Y, Nakagawa, A. | | Deposit date: | 2011-03-19 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity
J.Biol.Chem., 286, 2011
|
|
3AXA
 
 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
1UKL
 
 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
1UMK
 
 | | The Structure of Human Erythrocyte NADH-cytochrome b5 Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bando, S, Takano, T, Yubisui, T, Shirabe, K, Takeshita, M, Horii, C, Nakagawa, A. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human erythrocyte NADH-cytochrome b5 reductase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VCL
 
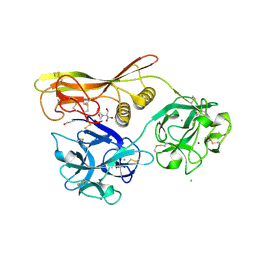 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
1WP1
 
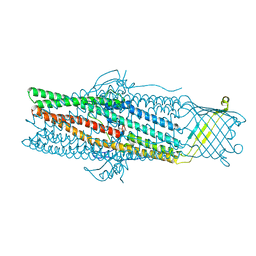 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
