3G02
 
 | |
3G0I
 
 | |
3MA0
 
 | |
3M9X
 
 | |
3M9W
 
 | |
3M1P
 
 | |
1RKD
 
 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1997-11-29 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 A resolution: insights into a new family of kinase structures.
Structure, 6, 1998
|
|
4AIC
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin, manganese and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose- 5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2WGS
 
 | | Crystal structure of Mycobacterium Tuberculosis Glutamine Synthetase in complex with a purine analogue inhibitor. | | Descriptor: | 1-(3,4-dichlorobenzyl)-3,7-dimethyl-8-morpholin-4-yl-3,7-dihydro-1H-purine-2,6-dione, CHLORIDE ION, GLUTAMINE SYNTHETASE 1 | | Authors: | Nilsson, M.T, Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterium Tuberculosis Glutamine Synthetase by Novel ATP-Competitive Inhibitors.
J.Mol.Biol., 393, 2009
|
|
2WHI
 
 | | Crystal structure of Mycobacterium Tuberculosis Glutamine Synthetase in complex with a purine analogue inhibitor and L-methionine-S- sulfoximine phosphate. | | Descriptor: | 1-(3,4-dichlorobenzyl)-3,7-dimethyl-8-morpholin-4-yl-3,7-dihydro-1H-purine-2,6-dione, CHLORIDE ION, GLUTAMINE SYNTHETASE 1, ... | | Authors: | Nilsson, M.T, Krajewski, W.W, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2009-05-05 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Inhibition of Mycobacterium Tuberculosis Glutamine Synthetase by Novel ATP-Competitive Inhibitors.
J.Mol.Biol., 393, 2009
|
|
2JD0
 
 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JCV
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JD2
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JCY
 
 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, SULFATE ION | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JCX
 
 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JD1
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with manganese and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, MANGANESE (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
1H46
 
 | | The catalytic module of Cel7D from Phanerochaete chrysosporium as a chiral selector: Structural studies of its complex with the b-blocker (R)-propranolol | | Descriptor: | (1E,2R)-1-(ISOPROPYLIMINO)-3-(1-NAPHTHYLOXY)PROPAN-2-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose, EXOGLUCANASE I | | Authors: | Munoz, I.G, Mowbray, S.L, Stahlberg, J. | | Deposit date: | 2002-10-03 | | Release date: | 2003-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The Catalytic Module of Cel7D from Phanerochaete Chrysosporium as a Chiral Selector: Structural Studies of its Complex with the Beta Blocker (R)-Propranolol
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1GUB
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, NICKEL (II) ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
1NN4
 
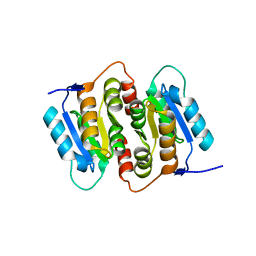 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
1MPD
 
 | |
1MPB
 
 | |
1MPC
 
 | |
1DPP
 
 | | DIPEPTIDE BINDING PROTEIN COMPLEX WITH GLYCYL-L-LEUCINE | | Descriptor: | DIPEPTIDE BINDING PROTEIN, GLYCINE, LEUCINE | | Authors: | Dunten, P, Mowbray, S.L. | | Deposit date: | 1995-08-11 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the dipeptide binding protein from Escherichia coli involved in active transport and chemotaxis.
Protein Sci., 4, 1995
|
|
1GPI
 
 | |
