3TZX
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in tetragonal apo form at 2.3 A | | Descriptor: | 12-mer peptide, GLYCEROL, Polyketide synthase PKS13, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
3TTQ
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3UEQ
 
 | | Crystal structure of amylosucrase from Neisseria polysaccharea in complex with turanose | | Descriptor: | 3-O-alpha-D-glucopyranosyl-D-fructose, Amylosucrase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guerin, F, Pizzut-Serin, S, Potocki-Veronese, G, Guillet, V, Mourey, L, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
3UER
 
 | | Crystal structure of amylosucrase from Deinococcus geothermalis in complex with turanose | | Descriptor: | Amylosucrase, alpha-D-glucopyranose-(1-3)-alpha-D-fructofuranose, alpha-D-glucopyranose-(1-3)-beta-D-fructofuranose | | Authors: | Guerin, F, Pizzut-Serin, S, Potocki-Veronese, G, Guillet, V, Mourey, L, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
3UCQ
 
 | | Crystal structure of amylosucrase from Deinococcus geothermalis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL | | Authors: | Guerin, F, Pizzut-Serin, S, Guillet, V, Mourey, L, Potocki-Veronese, G, Remaud-Simeon, M, Andre, I, Tranier, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Investigation of the Thermostability and Product Specificity of Amylosucrase from the Bacterium Deinococcus geothermalis.
J.Biol.Chem., 287, 2012
|
|
6QYG
 
 | |
6QYF
 
 | |
6QWU
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 5.5 with Mn2+ and CoA. | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
6QXQ
 
 | |
6QXR
 
 | | 4'-phosphopantetheinyl transferase PptAb from Mycobacterium abscessus at pH 8.5 with Mn2+ and CoA. | | Descriptor: | CALCIUM ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
4FLQ
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290I from Neisseria polysaccharea. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLO
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290C from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
4FLS
 
 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
6RCX
 
 | | Mycobacterial 4'-phosphopantetheinyl transferase PptAb in complex with the ACP domain of PpsC. | | Descriptor: | CACODYLATE ION, COENZYME A, MANGANESE (II) ION, ... | | Authors: | Nguyen, M.C, Mourey, L, Pedelacq, J.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4'-phosphopantetheinyl transferases.
Febs J., 287, 2020
|
|
4FLR
 
 | | Crystal structure of Amylosucrase double mutant A289P-F290L from Neisseria polysaccharea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amylosucrase, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
7NBU
 
 | | Structure of the HigB1 toxin mutant K95A from Mycobacterium tuberculosis (Rv1955) and its target, the cspA mRNA, on the E. coli Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Giudice, E, Mansour, M, Chat, S, D'Urso, G, Gillet, R, Genevaux, P. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Substrate recognition and cryo-EM structure of the ribosome-bound TAC toxin of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
1AGJ
 
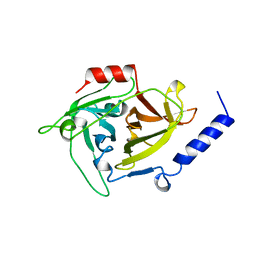 | | EPIDERMOLYTIC TOXIN A FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | EPIDERMOLYTIC TOXIN A | | Authors: | Cavarelli, J. | | Deposit date: | 1997-03-25 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of Staphylococcus aureus epidermolytic toxin A, an atypic serine protease, at 1.7 A resolution.
Structure, 5, 1997
|
|
8AJ2
 
 | |
5J1B
 
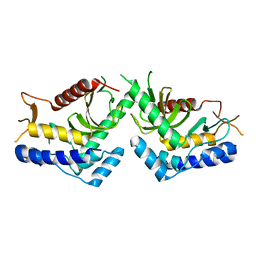 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - WT. | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Maveyraud, L, Batista, M, Martin-yken, H, Francois, J.M, Zerbib, D, Mourey, L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-04-05 | | Last modified: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biophysical characterization of Knr4, an intrinsically disordered hub protein form Saccharomyces cerevisiae
To Be Published
|
|
2JTG
 
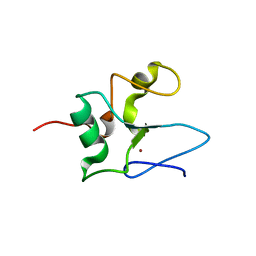 | | Solution structure of the THAP-zinc finger of THAP1 | | Descriptor: | THAP domain-containing protein 1, ZINC ION | | Authors: | Bessiere, D, Campagne, S, Milon, A, Gervais, V. | | Deposit date: | 2007-07-31 | | Release date: | 2007-12-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the THAP zinc finger of THAP1, a large C2CH DNA-binding module linked to Rb/E2F pathways
J.Biol.Chem., 283, 2008
|
|
1FQW
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
