2PD4
 
 | | Crystal Structure of the Helicobacter pylori Enoyl-Acyl Carrier Protein Reductase in Complex with Hydroxydiphenyl Ether Compounds, Triclosan and Diclosan | | Descriptor: | DICLOSAN, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, H.H, Moon, J.H, Suh, S.W. | | Deposit date: | 2007-03-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Helicobacter pylori enoyl-acyl carrier protein reductase in complex with hydroxydiphenyl ether compounds, triclosan and diclosan
Proteins, 69, 2007
|
|
5F1G
 
 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5GZW
 
 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
1FA2
 
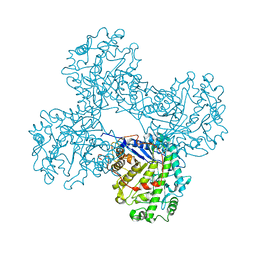 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
6AIL
 
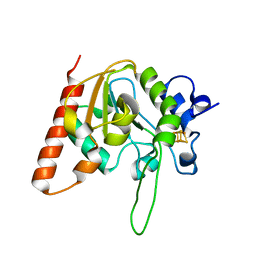 | | CRYSTAL STRUCTURE AT 1.3 ANGSTROMS RESOLUTION OF A NOVEL UDG, UdgX, FROM Mycobacterium smegmatis | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase X | | Authors: | Ahn, W.C, Aroli, S, Varshney, V, Woo, E.J. | | Deposit date: | 2018-08-24 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.335 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJR
 
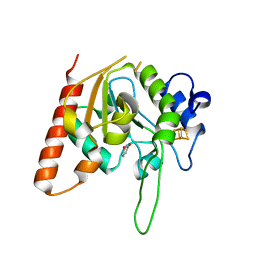 | | Complex form of Uracil DNA glycosylase X and uracil | | Descriptor: | IRON/SULFUR CLUSTER, URACIL, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.341 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJO
 
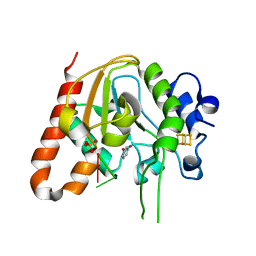 | | Complex form of Uracil DNA glycosylase X and uracil-DNA. | | Descriptor: | DNA (5'-D(P*(ORP)P*TP*T)-3'), IRON/SULFUR CLUSTER, PHOSPHATE ION, ... | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJP
 
 | | Complex form of Uracil DNA glycosylase X and deoxyuridine monophosphate. | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJQ
 
 | | E52Q mutant form of Uracil DNA glycosylase X from Mycobacterium smegmatis. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.342 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
6AJS
 
 | | H109S mutant form of Uracil DNA glycosylase X. | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Ahn, W.C, Aroli, S, Varshney, U, Woo, E.J. | | Deposit date: | 2018-08-28 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Covalent binding of uracil DNA glycosylase UdgX to abasic DNA upon uracil excision.
Nat.Chem.Biol., 15, 2019
|
|
2OKL
 
 | |
