3CRO
 
 | |
2CRO
 
 | |
1D6M
 
 | |
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | REPRESSOR PROTEIN CI | | 著者 | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | 登録日 | 1988-12-08 | | 公開日 | 1989-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
5HM5
 
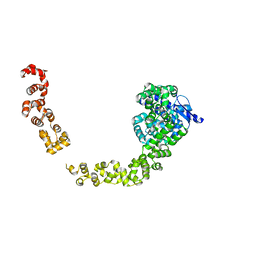 | |
4WLW
 
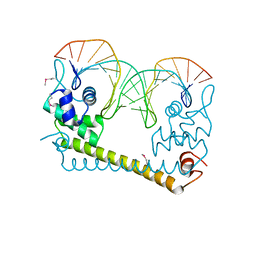 | | CRYSTAL STRUCTURE OF THE AG(I) (ACTIVATOR) FORM OF E. COLI CUER, A COPPER EFFLUX REGULATOR, BOUND TO COPA PROMOTER DNA | | 分子名称: | DNA NON-TEMPLATE STRAND (5-D(*DGP*DAP*DCP*DCP *DTP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DGP*DCP*DTP*DGP*DGP*DAP *DAP*DGP*DGP*DTP*DC)-3, DNA TEMPLATE STRAND (5-D(*DGP*DAP*DCP*DCP*DTP *DTP*DCP*DCP*DAP*DGP*DCP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DAP *DGP*DGP*DTP*DC)-3, HTH-type transcriptional regulator CueR, ... | | 著者 | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | 登録日 | 2014-10-08 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | TRANSCRIPTION. Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
4WLS
 
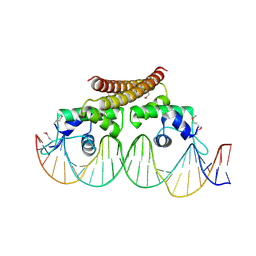 | | Crystal structure of the metal-free (repressor) form of E. Coli CUER, a copper efflux regulator, bound to COPA promoter DNA | | 分子名称: | COPA PROMOTER DNA NON-TEMPLATE STRAND, COPA PROMOTER DNA NON-TEMPLATE STRAND (ALTERNATE CONFORMATION), COPA PROMOTER DNA TEMPLATE STRAND, ... | | 著者 | Philips, S.J, Canalizo-Hernandez, M, Mondragon, A, O'Halloran, T.V. | | 登録日 | 2014-10-08 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.105 Å) | | 主引用文献 | Allosteric transcriptional regulation via changes in the overall topology of the core promoter.
Science, 349, 2015
|
|
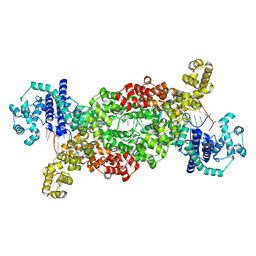
8DF7
 
 | |
8DFB
 
 | |
8DF9
 
 | |
8DF8
 
 | |
2F4Q
 
 | |
4ZQA
 
 | |
7LJP
 
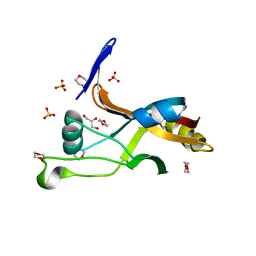 | | Structure of Thermotoga maritima SmpB | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, SULFATE ION, ... | | 著者 | Chan, C.W, Mondragon, A. | | 登録日 | 2021-01-29 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of Thermotoga maritima SmpB reveals its C-terminal tail domain in a helical conformation mimicking that of a ribosome-bound state
To Be Published
|
|
6N1Q
 
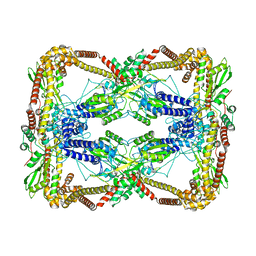 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | 分子名称: | DNA gyrase subunit A | | 著者 | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | 登録日 | 2018-11-10 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (5.16 Å) | | 主引用文献 | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6NBU
 
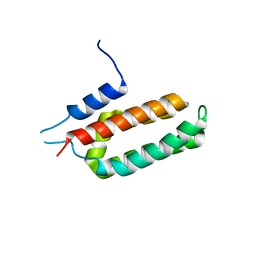 | |
6NBT
 
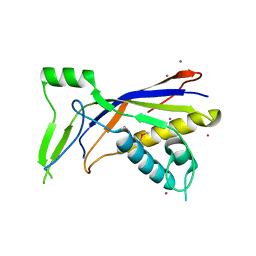 | |
4MTE
 
 | |
1I9S
 
 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | 分子名称: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | 著者 | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | 登録日 | 2001-03-20 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
1I9T
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | 分子名称: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | 著者 | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | 登録日 | 2001-03-20 | | 公開日 | 2001-05-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
7UZX
 
 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | 分子名称: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V00
 
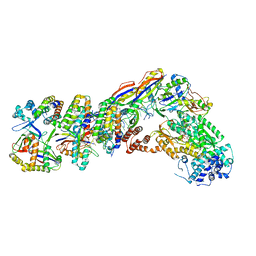 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZZ
 
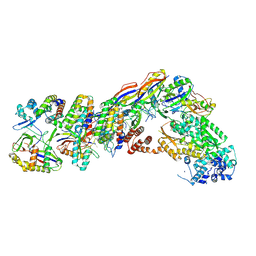 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.45 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZY
 
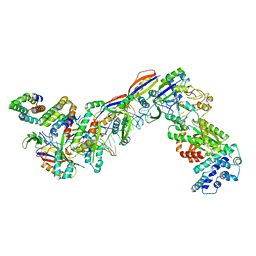 | | Staphylococcus epidermidis RP62A CRISPR effector complex with non-self target RNA 2 | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7V02
 
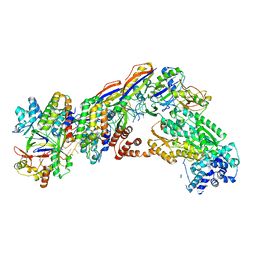 | | Staphylococcus epidermidis RP62A CRISPR short effector complex | | 分子名称: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | 著者 | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | 登録日 | 2022-05-09 | | 公開日 | 2022-07-06 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (4.97 Å) | | 主引用文献 | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
