5YN2
 
 | | Crystal structure of apo Pullulanase from Klebsiella pneumoniae in space group P43212 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PulA protein | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YN7
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 0.1 mM beta-cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YND
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DUF3372 domain-containing protein, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YNH
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5YYP
 
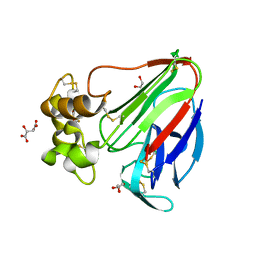 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
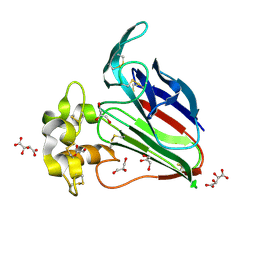 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5Z6B
 
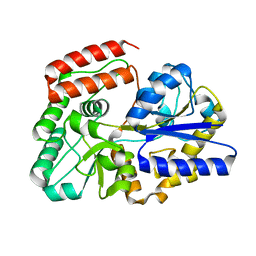 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | Authors: | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
5YYR
 
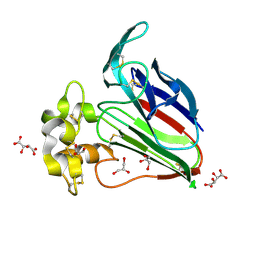 | | Structure K106A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Ohta, K, Mitsumoto, M, Mikami, B, Suzuki, M, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5Z6C
 
 | |
6A4U
 
 | | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ferritin, ... | | Authors: | Masuda, T, Mikami, B, Zang, J, Zhao, G. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The first crystal structure of crustacean ferritin that is a hybrid type of H and L ferritin
Protein Sci., 27, 2018
|
|
2Z42
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2007-06-12 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Recognition by Family 7 Alginagte Lyase from Sphingomonas sp. A1
To be published
|
|
2ZAB
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Cmplex with Product (GGG) | | Descriptor: | Alginate lyase, GLYCEROL, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2Z9W
 
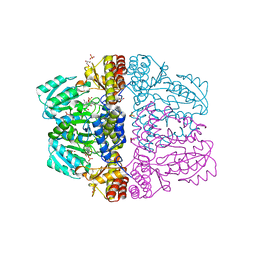 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxal | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZAC
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' Y284F in Complex with Product (MMG) | | Descriptor: | Alginate lyase, GLYCEROL, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2ZUY
 
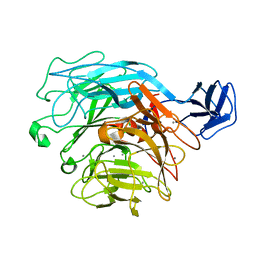 | | Crystal structure of exotype rhamnogalacturonan lyase YesX | | Descriptor: | CALCIUM ION, YesX protein | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
2Z72
 
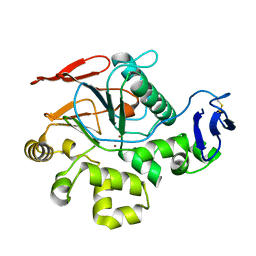 | | New Structure Of Cold-Active Protein Tyrosine Phosphatase At 1.1 Angstrom | | Descriptor: | Protein-tyrosine-phosphatase, ZINC ION | | Authors: | Tsuruta, H, Mikami, B, Yamamoto, C, Yamagata, H. | | Deposit date: | 2007-08-10 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The role of group bulkiness in the catalytic activity of psychrophile cold-active protein tyrosine phosphatase
Febs J., 275, 2008
|
|
2Z9X
 
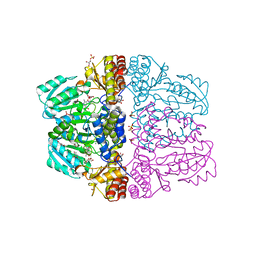 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZBL
 
 | | Functional annotation of Salmonella enterica yihS-encoded protein | | Descriptor: | Putative isomerase, beta-D-mannopyranose | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-24 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
2ZA9
 
 | | Crystal Structure of Alginate lyase A1-II' N141C/N199C | | Descriptor: | Alginate lyase, SULFATE ION | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2Z9U
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti at 2.0 A resolution | | Descriptor: | Aspartate aminotransferase, GLYCEROL, SULFATE ION | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZUX
 
 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein, ... | | Authors: | Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2008-10-28 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural determinants responsible for substrate recognition and mode of action in family 11 polysaccharide lyases
J.Biol.Chem., 284, 2009
|
|
2Z8R
 
 | | Crystal structure of rhamnogalacturonan lyase YesW at 1.40 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, YesW protein | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
2Z9V
 
 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxamine | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Aspartate aminotransferase, GLYCEROL, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
2ZAA
 
 | | Crystal Structure of Family 7 Alginate Lyase A1-II' H191N/Y284F in Complex with Substrate (GGMG) | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase, GLYCEROL | | Authors: | Ogura, K, Yamasaki, M, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-02 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate Recognition in Tunnel of Family 7 Alginate Lyase from Sphingomonas sp. A1
To be Published
|
|
2Z8S
 
 | | Crystal structure of rhamnogalacturonan lyase YesW complexed with digalacturonic acid | | Descriptor: | CALCIUM ION, YesW protein, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Ochiai, A, Itoh, T, Maruyama, Y, Kawamata, A, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Structural Fold in Polysaccharide Lyases: BACILLUS SUBTILIS FAMILY 11 RHAMNOGALACTURONAN LYASE YesW WITH AN EIGHT-BLADED -PROPELLER
J.Biol.Chem., 282, 2007
|
|
