1W8I
 
 | | The Structure of gene product af1683 from Archaeoglobus fulgidus. | | Descriptor: | PUTATIVE VAPC RIBONUCLEASE AF_1683 | | Authors: | Midwest Center for Structural Genomics (MCSG), Cuff, M.E, Zhang, R, Ginell, S.L, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of Gene Product Af1683 from Archaeoglobus Fulgidus
To be Published
|
|
1YQH
 
 | |
2G17
 
 | |
6OK3
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.353 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ONW
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-22 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORC
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6ORK
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
7M5H
 
 | | Crystal structure of conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase | | Authors: | Nocek, B, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-03-23 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of conserved protein from Enterococcus faecalis V583
To Be Published
|
|
4DWJ
 
 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate | | Descriptor: | THYMIDINE-5'-PHOSPHATE, Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in complex with Thymidine Monophosphate
To be Published
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
4YF1
 
 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
4MNU
 
 | | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from Listeria monocytogenes | | Descriptor: | GLYCEROL, SULFATE ION, SlyA-like transcription regulator | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal Structure of Uncharacterized SlyA-like Transcription Regulator from
Listeria monocytogenes
To be Published
|
|
4N01
 
 | |
4MK6
 
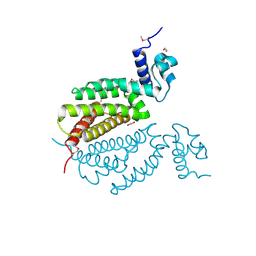 | | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e | | Descriptor: | 1,2-ETHANEDIOL, Probable Dihydroxyacetone Kinase Regulator DHSK_reg | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-04 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Probable Dihydroxyacetone Kinase Regulator DHSK_reg from Listeria monocytogenes EGD-e
To be Published
|
|
4MO9
 
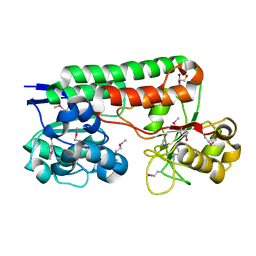 | | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula | | Descriptor: | GLYCEROL, Periplasmic binding protein, trimethylamine oxide | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal Structure of TroA-like Periplasmic Binding Protein FepB from Veillonella parvula
To be Published
|
|
4N04
 
 | |
4MNR
 
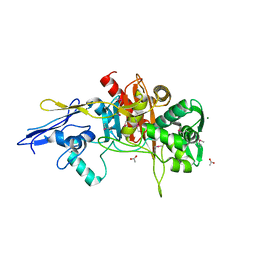 | | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Peptidoglycan glycosyltransferase | | Authors: | Kim, Y, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Crystal Structure of D,D-Transpeptidase Domain of Peptidoglycan Glycosyltransferase from Eggerthella lenta
To be Published
|
|
4MX8
 
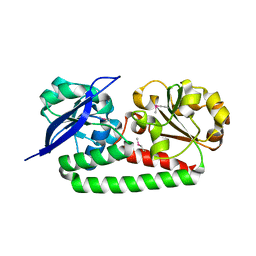 | |
4RIT
 
 | | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-10-07 | | Release date: | 2014-10-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The yellow crystal structure of pyridoxal-dependent decarboxylase from sphaerobacter thermophilus dsm 20745
To be Published
|
|
4RAW
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Ampicillin
To be Published
|
|
4RGP
 
 | | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans | | Descriptor: | CALCIUM ION, Csm6_III-A, GLYCEROL, ... | | Authors: | Kim, Y, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal Structure of Uncharacterized CRISPR/Cas System-associated Protein Csm6 from Streptococcus mutans
To be Published
|
|
4RIZ
 
 | |
4RJ0
 
 | |
4S1A
 
 | | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC 27405 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, TETRAETHYLENE GLYCOL, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-09 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a hypothetical protein Cthe_0052 from Ruminiclostridium thermocellum ATCC27405
To be Published
|
|
4RM1
 
 | |
