1XFR
 
 | |
7QNP
 
 | | Designed Armadillo repeat protein N(A4)M4C(AII) co-crystallized with hen egg white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Designed Armadillo Repeat Protein N(A4)M4C(AII), ... | | Authors: | Michel, E, Mittl, P.R.E, Plueckthun, A. | | Deposit date: | 2021-12-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Improved Repeat Protein Stability by Combined Consensus and Computational Protein Design.
Biochemistry, 62, 2023
|
|
6FBL
 
 | | NMR Solution Structure of MINA-1(254-334) | | Descriptor: | MINA-1 | | Authors: | Michel, E, Allain, F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans.
Cell Death Differ., 26, 2019
|
|
6G04
 
 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6G03
 
 | | NMR Solution Structure of yeast TSR2(1-152) | | Descriptor: | Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
2MFF
 
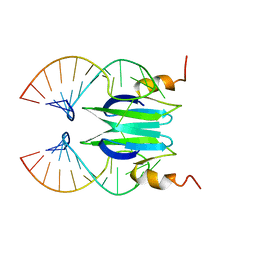 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
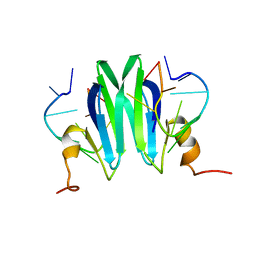 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFE
 
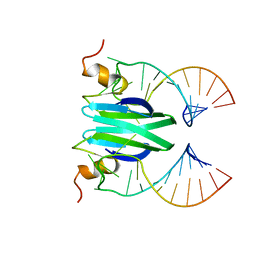 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFG
 
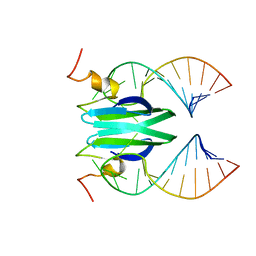 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFC
 
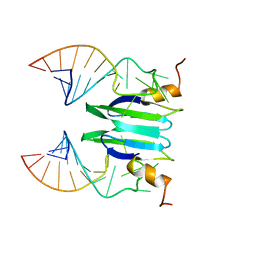 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MF0
 
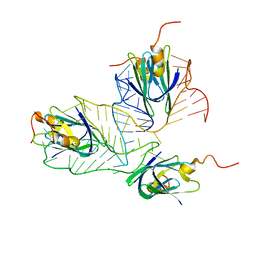 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
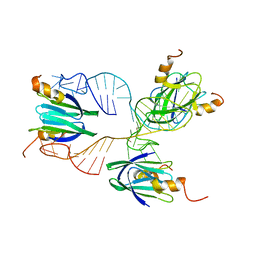 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
7R0R
 
 | |
8OH7
 
 | |
8RDY
 
 | | Saccharomyces cerevisiae Prp43 helicase in complex with Pxr1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione S-transferase class-mu 26 kDa isozyme,Protein PXR1, MAGNESIUM ION, ... | | Authors: | Rabl, J, Portugal Calisto, D, Panse, V.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | An inhibitory segment within G-patch activators tunes Prp43-ATPase activity during ribosome assembly.
Nat Commun, 15, 2024
|
|
3P7B
 
 | | p38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(5-oxo-1,4-diazepan-1-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P7A
 
 | | p38 inhibitor-bound | | Descriptor: | 1-[5-tert-butyl-2-(1,1-dioxidothiomorpholin-4-yl)thiophen-3-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P79
 
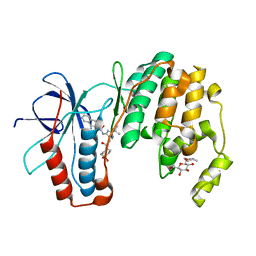 | | P38 inhibitor-bound | | Descriptor: | 1-{3-tert-butyl-1-[2-(1,1-dioxidothiomorpholin-4-yl)-2-oxoethyl]-1H-pyrazol-5-yl}-3-naphthalen-2-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P7C
 
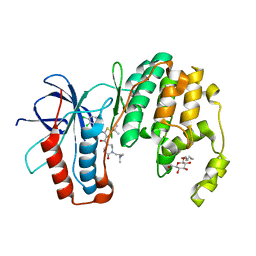 | | p38 inhibitor-bound | | Descriptor: | 1-[5-tert-butyl-3-({4-[2-(dimethylamino)ethyl]-5-oxo-1,4-diazepan-1-yl}carbonyl)thiophen-2-yl]-3-(2,3-dichlorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P78
 
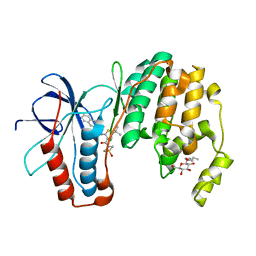 | | P38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(1,1-dioxidothiomorpholin-4-yl)carbonyl]thiophen-2-yl}-3-naphthalen-2-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3P5K
 
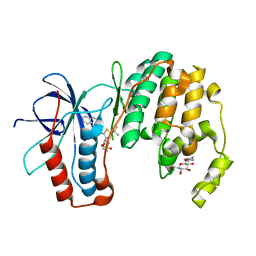 | | P38 inhibitor-bound | | Descriptor: | 1-{5-tert-butyl-3-[(1,1-dioxidothiomorpholin-4-yl)carbonyl]thiophen-2-yl}-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H. | | Deposit date: | 2010-10-08 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4QH3
 
 | |
4QGZ
 
 | |
