3BBD
 
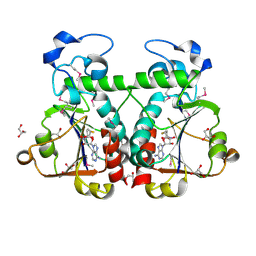 | | M. jannaschii Nep1 complexed with S-adenosyl-homocysteine | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBE
 
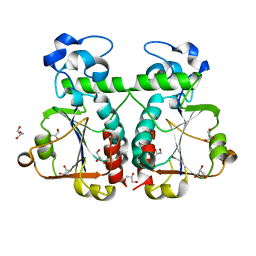 | | M. jannaschii Nep1 | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
3BBH
 
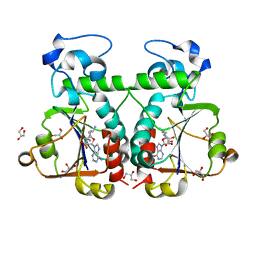 | | M. jannaschii Nep1 complexed with Sinefungin | | Descriptor: | GLYCEROL, Ribosome biogenesis protein NEP1-like, SINEFUNGIN | | Authors: | Taylor, A.B, Meyer, B, Leal, B.Z, Kotter, P, Hart, P.J, Entian, K.-D, Wohnert, J. | | Deposit date: | 2007-11-09 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of Nep1 reveals an extended SPOUT-class methyltransferase fold and a pre-organized SAM-binding site.
Nucleic Acids Res., 36, 2008
|
|
2JNR
 
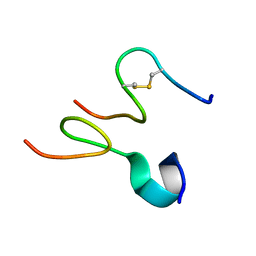 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
3ZDV
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(2,4,6-trimethylphenylsulfonylamido)-6-deoxy-alpha-D-mannopyranoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trimethylbenzenesulfonamide, CALCIUM ION, ... | | Authors: | Hauck, D, Joachim, I, Frommeyer, B, Varrot, A, Philipp, B, MOller, H.M, Imberty, A, Exner, T.E, Titz, A. | | Deposit date: | 2012-11-30 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Discovery of Two Classes of Potent Glycomimetic Inhibitors of Pseudomonas Aeruginosa Lecb with Distinct Binding Modes.
Acs Chem.Biol., 8, 2013
|
|
4ES8
 
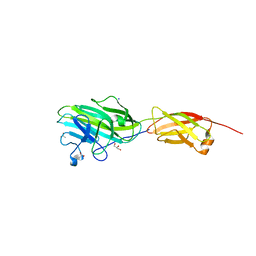 | | Crystal Structure of the adhesin domain of Epf from Streptococcus pyogenes in P212121 | | Descriptor: | ACETATE ION, Epf, GLYCEROL, ... | | Authors: | Linke, C, Siemens, N, Kreikemeyer, B, Baker, E.N. | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Extracellular Protein Factor Epf from Streptococcus pyogenes Is a Cell Surface Adhesin That Binds to Cells through an N-terminal Domain Containing a Carbohydrate-binding Module.
J.Biol.Chem., 287, 2012
|
|
4ES9
 
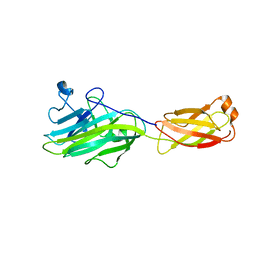 | |
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7L5D
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with demethylated analog of masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
2L6S
 
 | | Efficacy of an HIV-1 entry inhibitor targeting the GP41 fusion peptide | | Descriptor: | VIR-576 | | Authors: | Forssmann, W, The, Y, Stoll, M, Adermann, K, Albrecht, U, Barlos, K, Busmann, A, Canales-Mayordomo, A, Gimenez-Gallego, G, Hirsch, J, Jimenez-Barbero, J, Meyer-Olson, D, Muench, J, Perez-Castells, J, Standker, L, Kirchhoff, F, Schmidt, R.E. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide.
Sci Transl Med, 2, 2010
|
|
2L6T
 
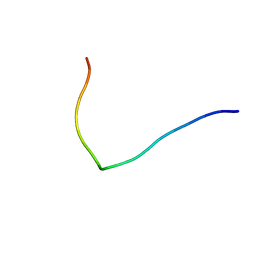 | | Efficacy of an HIV-1 entry inhibitor targeting the GP41 fusion peptide | | Descriptor: | VIR-576 | | Authors: | Forssmann, W, The, Y, Stoll, M, Adermann, K, Albrecht, U, Barlos, K, Busmann, A, Can les-Mayordomo, A, Gimenez-Gallego, G, Hirsch, J, Jimenez-Barbero, J, Meyer-Olson, D, Muench, J, Perez-Castells, J, Staendker, L, Kirchhoff, F, Schmidt, R.E. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide.
Sci Transl Med, 2, 2010
|
|
7BH9
 
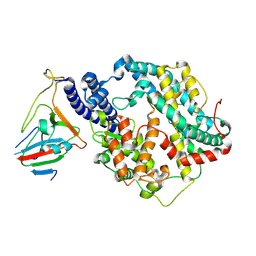 | | SARS-CoV-2 RBD-62 in complex with ACE2 peptidase domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Elad, N, Dym, O, Zahradnik, J, Schreiber, G. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-17 | | Last modified: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution.
Nat Microbiol, 6, 2021
|
|
5APG
 
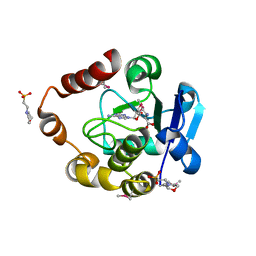 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5AP8
 
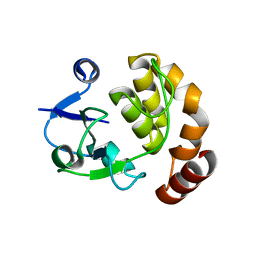 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
8OU1
 
 | |
8K6T
 
 | | The minor pilin structure of FctB3 in Streptococcus | | Descriptor: | FctB3, GLYCEROL | | Authors: | Takebe, K, Sangawa, T, Suzuki, M, Nakata, M. | | Deposit date: | 2023-07-25 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of FctB3 crystal structure and insight into its structural stabilization and pilin linkage mechanisms.
Arch.Microbiol., 206, 2023
|
|
7PKJ
 
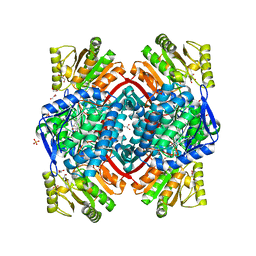 | | Streptococcus pyogenes apo GapN | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
7PKC
 
 | | Streptococcus pyogenes Apo-GapN C284S variant | | Descriptor: | GLYCEROL, Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Schindelin, H, Albert, L. | | Deposit date: | 2021-08-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Non-phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase GapN Is a Potential New Drug Target in Streptococcus pyogenes.
Front Microbiol, 13, 2022
|
|
6G2G
 
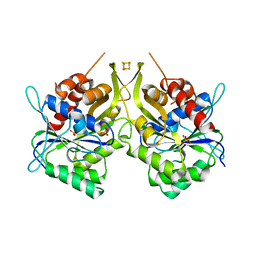 | | Fe-S assembly Cfd1 | | Descriptor: | Cytosolic Fe-S cluster assembly factor CFD1, IRON/SULFUR CLUSTER, SULFATE ION | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2018-03-23 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Function and crystal structure of the dimeric P-loop ATPase CFD1 coordinating an exposed [4Fe-4S] cluster for transfer to apoproteins.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
