2P5C
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2PCH
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2OWD
 
 | | Crystal structure of TTHB049 from Thermus thermophilus HB8 | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, GLYCEROL, SODIUM ION | | Authors: | Sugahara, M, Taketa, M, Ono, N, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-16 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of TTHB049 from Thermus thermophilus HB8
To be Published
|
|
2P2X
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sugahara, M, Ono, N, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-08 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
3TPM
 
 | |
3TPQ
 
 | |
3TPO
 
 | |
3ULB
 
 | |
3ULC
 
 | |
3VOQ
 
 | |
3VYC
 
 | |
3W3X
 
 | | Crystal structure of Kap121p bound to Pho4p | | Descriptor: | Importin subunit beta-3, Phosphate system positive regulatory protein PHO4 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3Y
 
 | | Crystal structure of Kap121p bound to Nup53p | | Descriptor: | Importin subunit beta-3, Nucleoporin NUP53 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3T
 
 | | Crystal structure of Kap121p | | Descriptor: | Importin subunit beta-3 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3V
 
 | |
3W3W
 
 | | Crystal structure of Kap121p bound to Ste12p | | Descriptor: | Importin subunit beta-3, Protein STE12 | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3Z
 
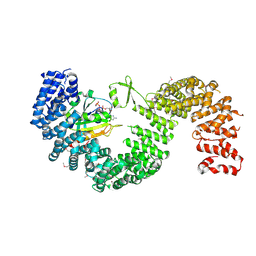 | | Crystal structure of Kap121p bound to RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin subunit beta-3, ... | | Authors: | Kobayashi, J, Matsuura, Y. | | Deposit date: | 2012-12-28 | | Release date: | 2013-04-10 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cell-cycle-dependent nuclear import mediated by the karyopherin Kap121p.
J.Mol.Biol., 425, 2013
|
|
3W3U
 
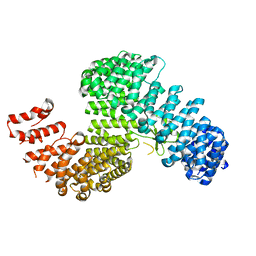 | |
2EK4
 
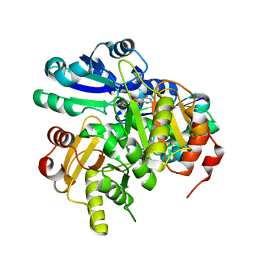 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L8M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Shimada, H, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L8M)
To be Published
|
|
2EK3
 
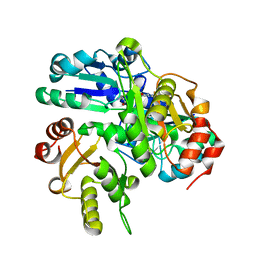 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L3M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Shimada, H, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L3M)
To be Published
|
|
2EL3
 
 | | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L242M) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, diphthine synthase | | Authors: | Asada, Y, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural study of Project ID PH0725 from Pyrococcus horikoshii OT3 (L242M)
To be Published
|
|
3X3U
 
 | |
3WYF
 
 | | Crystal structure of Xpo1p-Yrb2p-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GUANOSINE-5'-TRIPHOSPHATE, Gsp1p, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Cell Rep, 9, 2014
|
|
2EKZ
 
 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L52M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase | | Authors: | Asada, Y, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-26 | | Release date: | 2007-10-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L52M)
To be Published
|
|
3WYG
 
 | | Crystal structure of Xpo1p-PKI-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GUANOSINE-5'-TRIPHOSPHATE, Gsp1p, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Cell Rep, 9, 2014
|
|
