2KGF
 
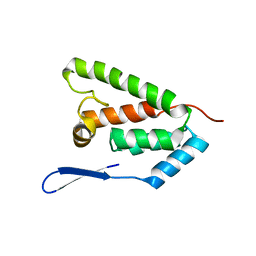 | | N-terminal domain of capsid protein from the Mason-Pfizer monkey virus | | Descriptor: | Capsid protein p27 | | Authors: | Macek, P, Chmelik, J, Zidek, L, Kaderavek, P, Padrta, P, Ruml, T, Pichova, I, Rumlova, M, Sklenar, V. | | Deposit date: | 2009-03-10 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of capsid protein from the mason-pfizer monkey virus
J.Mol.Biol., 392, 2009
|
|
8B7J
 
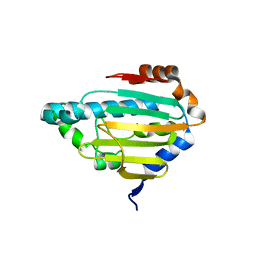 | | Human HSP90 alpha ATP Binding Domain, ATP-lid closed conformation, R46A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brustcher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
8B7I
 
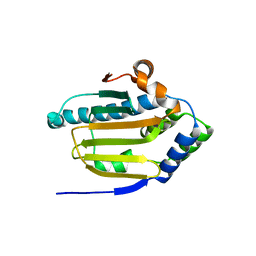 | | Human HSP90 alpha ATP Binding Domain, ATP-lid open conformation, R60A | | Descriptor: | HSP90AA1 protein | | Authors: | Rioual, E, Henot, F, Favier, A, Macek, P, Crublet, E, Josso, P, Brutscher, B, Frech, M, Gans, P, Loison, C, Boisbouvier, J. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Visualizing the transiently populated closed-state of human HSP90 ATP binding domain.
Nat Commun, 13, 2022
|
|
1IAZ
 
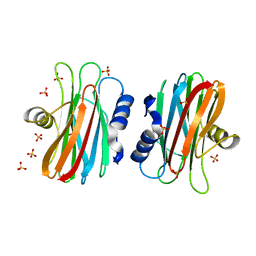 | | EQUINATOXIN II | | Descriptor: | EQUINATOXIN II, SULFATE ION | | Authors: | Athanasiadis, A, Anderluh, G, Macek, P, Turk, D. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the soluble form of equinatoxin II, a pore-forming toxin from the sea anemone Actinia equina.
Structure, 9, 2001
|
|
6F3K
 
 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
6SJW
 
 | | Structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
6SJX
 
 | | Precursor structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
5CVW
 
 | | CRYSTAL STRUCTURE OF RTX DOMAIN BLOCK V OF ADENYLATE CYCLASE TOXIN FROM BORDETELLA PERTUSSIS | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional hemolysin/adenylate cyclase, CALCIUM ION, ... | | Authors: | Motlova, L, Barinka, C, Bumba, L. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Calcium-Driven Folding of RTX Domain beta-Rolls Ratchets Translocation of RTX Proteins through Type I Secretion Ducts.
Mol.Cell, 62, 2016
|
|
5CXL
 
 | |
1TZQ
 
 | | Crystal structure of the equinatoxin II 8-69 double cysteine mutant | | Descriptor: | Equinatoxin II | | Authors: | Kristan, K, Podlesek, Z, Hojnik, V, Gutirrez-Aguirre, I, Guncar, G, Turk, D.A, Gonzalez-Maas, J.M, Lakey, J.H, Anderluh, G. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pore formation by equinatoxin, a eukaryotic pore-forming toxin, requires a flexible N-terminal region and a stable beta-sandwich
J.Biol.Chem., 279, 2004
|
|
6ZC2
 
 | | Crystal structure of RahU protein in complex with TRIS molecule | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RahU protein | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structure of RahU, an aegerolysin protein from the human pathogen Pseudomonas aeruginosa, and its interaction with membrane ceramide phosphorylethanolamine.
Sci Rep, 11, 2021
|
|
6ZC1
 
 | |
6R8N
 
 | | STRUCTURE DETERMINATION OF THE TETRAHEDRAL AMINOPEPTIDASE TET2 FROM P. HORIKOSHII BY USE OF COMBINED SOLID-STATE NMR, SOLUTION-STATE NMR AND EM DATA 4.1 A, FOLLOWED BY REAL_SPACE_REFINEMENT AT 4.1 A | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Colletier, J.-P, Gauto, D, Estrozi, L, Favier, A, Effantin, G, Schoehn, G, Boisbouvier, J, Schanda, P. | | Deposit date: | 2019-04-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
