3CAU
 
 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | 分子名称: | 60 kDa chaperonin | | 著者 | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | 登録日 | 2008-02-20 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3C9V
 
 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | 分子名称: | 60 kDa chaperonin | | 著者 | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | 登録日 | 2008-02-18 | | 公開日 | 2008-09-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
5JUL
 
 | | Near atomic structure of the Dark apoptosome | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK | | 著者 | Cheng, T.C, Akey, I.V, Yuan, S, Yu, Z, Ludtke, S.J, Akey, C.W. | | 登録日 | 2016-05-10 | | 公開日 | 2017-02-22 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | A Near-Atomic Structure of the Dark Apoptosome Provides Insight into Assembly and Activation.
Structure, 25, 2017
|
|
6MU2
 
 | | Structure of full-length IP3R1 channel in the Apo-state | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
4V4L
 
 | | Structure of the Drosophila apoptosome | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK, MAGNESIUM ION | | 著者 | Yuan, S, Topf, M, Akey, C.W, Ludtke, S.J. | | 登録日 | 2010-10-04 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Structure of the Drosophila apoptosome at 6.9 angstrom resolution
Structure, 19, 2011
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | 著者 | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | 登録日 | 2013-06-17 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
1TT9
 
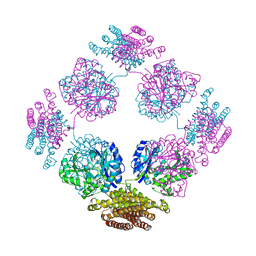 | | Structure of the bifunctional and Golgi associated formiminotransferase cyclodeaminase octamer | | 分子名称: | Formimidoyltransferase-cyclodeaminase (Formiminotransferase- cyclodeaminase) (FTCD) (58 kDa microtubule-binding protein) | | 著者 | Mao, Y, Vyas, N.K, Vyas, M.N, Chen, D.H, Ludtke, S.J, Chiu, W, Quiocho, F.A. | | 登録日 | 2004-06-22 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Structure of the bifunctional and Golgi-associated formiminotransferase cyclodeaminase octamer
Embo J., 23, 2004
|
|
8EAR
 
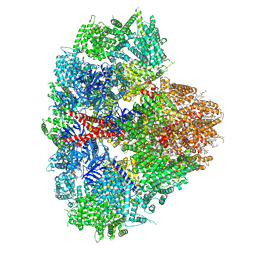 | | Structure of the full-length IP3R1 channel determined in the presence of Calcium/IP3/ATP | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | 著者 | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
8EAQ
 
 | | Structure of the full-length IP3R1 channel determined at high Ca2+ | | 分子名称: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 1, ... | | 著者 | Fan, G, Baker, M.R, Terry, L.E, Arige, V, Chen, M, Seryshev, A.B, Baker, M.L, Ludtke, S.J, Yule, D.I, Serysheva, I.I. | | 登録日 | 2022-08-29 | | 公開日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Conformational motions and ligand-binding underlying gating and regulation in IP 3 R channel.
Nat Commun, 13, 2022
|
|
6MU1
 
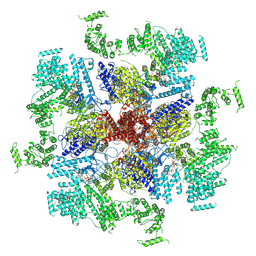 | | Structure of full-length IP3R1 channel bound with Adenophostin A | | 分子名称: | Adenophostin A, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | 登録日 | 2018-10-22 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-11-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | 分子名称: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | 著者 | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | 登録日 | 2021-06-17 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (37 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
3KTT
 
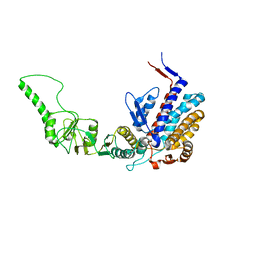 | | Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map | | 分子名称: | T-complex protein 1 subunit beta | | 著者 | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | 登録日 | 2009-11-25 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IYG
 
 | | Ca model of bovine TRiC/CCT derived from a 4.0 Angstrom cryo-EM map | | 分子名称: | T-complex protein 1 subunit, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | 著者 | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | 登録日 | 2009-11-28 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IYF
 
 | | Atomic Model of the Lidless Mm-cpn in the Open State | | 分子名称: | Chaperonin | | 著者 | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | 登録日 | 2009-10-23 | | 公開日 | 2010-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
3J7L
 
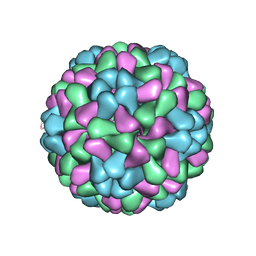 | | Full virus map of brome mosaic virus | | 分子名称: | Capsid protein | | 著者 | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | 登録日 | 2014-07-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
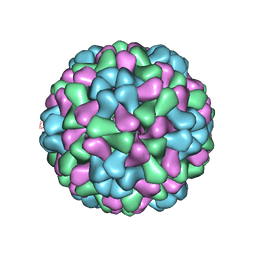 | | Virus model of brome mosaic virus (second half data set) | | 分子名称: | Capsid protein | | 著者 | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | 登録日 | 2014-07-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J45
 
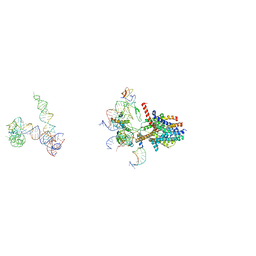 | | Structure of a non-translocating SecY protein channel with the 70S ribosome | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L23, 50S ribosomal protein L24, ... | | 著者 | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | 登録日 | 2013-06-18 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3J7M
 
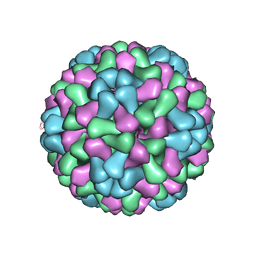 | | Virus model of brome mosaic virus (first half data set) | | 分子名称: | Capsid protein | | 著者 | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | 登録日 | 2014-07-18 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3JAV
 
 | | Structure of full-length IP3R1 channel in the apo-state determined by single particle cryo-EM | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Fan, G, Baker, M.L, Wang, Z, Baker, M.R, Sinyagovskiy, P.A, Chiu, W, Ludtke, S.J, Serysheva, I.I. | | 登録日 | 2015-06-30 | | 公開日 | 2015-10-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Gating machinery of InsP3R channels revealed by electron cryomicroscopy.
Nature, 527, 2015
|
|
3J46
 
 | | Structure of the SecY protein translocation channel in action | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L1, 50S ribosomal protein L23P, ... | | 著者 | Akey, C.W, Park, E, Menetret, J.F, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A. | | 登録日 | 2013-06-18 | | 公開日 | 2013-10-23 | | 最終更新日 | 2019-07-03 | | 実験手法 | ELECTRON MICROSCOPY (10.1 Å) | | 主引用文献 | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
3LOS
 
 | | Atomic Model of Mm-cpn in the Closed State | | 分子名称: | Chaperonin | | 著者 | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | 登録日 | 2010-02-04 | | 公開日 | 2010-03-16 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
6BG9
 
 | |
6PMO
 
 | |
6POM
 
 | | Cryo-EM structure of the full-length Bacillus subtilis glyQS T-box riboswitch in complex with tRNA-Gly | | 分子名称: | T-box GlyQS leader (155-MER), tRNAGly (75-MER) | | 著者 | Li, S, Su, Z, Zhang, J, Chiu, W. | | 登録日 | 2019-07-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | 分子名称: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | 著者 | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | 登録日 | 2021-06-13 | | 公開日 | 2022-01-26 | | 最終更新日 | 2022-02-02 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
