7OJT
 
 | | Crystal structure of unliganded PatA, a membrane associated acyltransferase from Mycobacterium smegmatis | | Descriptor: | GLYCEROL, Phosphatidylinositol mannoside acyltransferase | | Authors: | Anso, I, Wang, L, Marina, A, Paez-Perez, E.D, Perrone, S, Lowary, T.L, Trastoy, B, Guerin, M.E. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.67 Å) | | Cite: | Molecular ruler mechanism and interfacial catalysis of the integral membrane acyltransferase PatA.
Sci Adv, 7, 2021
|
|
4MO2
 
 | | Crystal Structure of UDP-N-acetylgalactopyranose mutase from Campylobacter jejuni | | Descriptor: | AMMONIUM ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Protsko, C, Poulin, M.B, Lowary, T.L, Sanders, D.A.R. | | Deposit date: | 2013-09-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity of a UDP-GalNAc Pyranose-Furanose Mutase: A Potential Therapeutic Target for Campylobacter jejuni Infections.
Chembiochem, 15, 2014
|
|
2YGM
 
 | | THE X-RAY CRYSTAL STRUCTURE OF TANDEM CBM51 MODULES OF SP3GH98, THE FAMILY 98 GLYCOSIDE HYDROLASE FROM STREPTOCOCCUS PNEUMONIAE SP3-BS71, IN COMPLEX WITH THE BLOOD GROUP B ANTIGEN | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION, SODIUM ION, ... | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
2YGL
 
 | | The X-ray crystal structure of tandem CBM51 modules of Sp3GH98, the family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 | | Descriptor: | BLOOD GROUP A-AND B-CLEAVING ENDO-BETA-GALACTOSIDASE, CALCIUM ION | | Authors: | Higgins, M.A, Ficko-Blean, E, Wright, C, Meloncelli, P.J, Lowary, T.L, Boraston, A.B. | | Deposit date: | 2011-04-18 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Overall Architecture and Receptor Binding of Pneumococcal Carbohydrate Antigen Hydrolyzing Enzymes.
J.Mol.Biol., 411, 2011
|
|
4FIY
 
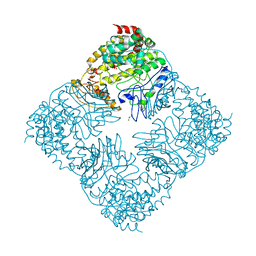 | | Crystal Structure of GlfT2 Complexed with UDP | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-galactofuranosyl transferase GlfT2, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
4FIX
 
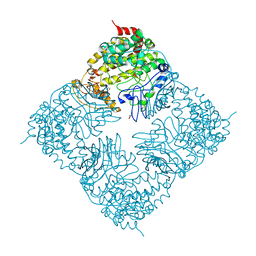 | | Crystal Structure of GlfT2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wheatley, R.W, Zheng, R.B, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2012-06-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan.
J.Biol.Chem., 287, 2012
|
|
6W98
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Acyl carrier protein, CALCIUM ION, ... | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-22 | | Release date: | 2020-05-13 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBX
 
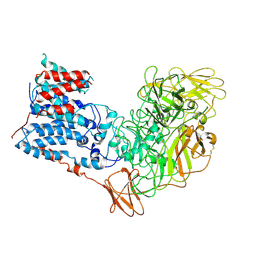 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 1 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6WBY
 
 | | Single-Particle Cryo-EM Structure of Arabinofuranosyltransferase AftD from Mycobacteria, Mutant R1389S Class 2 | | Descriptor: | CALCIUM ION, DUF3367 domain-containing protein | | Authors: | Tan, Y.Z, Zhang, L, Rodrigues, J, Zheng, R.B, Giacometti, S.I, Rosario, A.L, Kloss, B, Dandey, V.P, Wei, H, Brunton, R, Raczkowski, A.M, Athayde, D, Catalao, M.J, Pimentel, M, Clarke, O.B, Lowary, T.L, Archer, M, Niederweis, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structures and Regulation of Arabinofuranosyltransferase AftD from Mycobacteria.
Mol.Cell, 78, 2020
|
|
6X0O
 
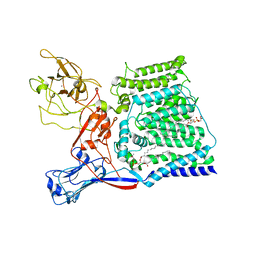 | | Single-Particle Cryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbB | | Authors: | Tan, Y.Z, Rodrigues, J, Keener, J.E, Zheng, R.B, Brunton, R, Kloss, B, Giacometti, S.I, Rosario, A.L, Zhang, L, Niederweis, M, Clarke, O.B, Lowary, T.L, Marty, M.T, Archer, M, Potter, C.S, Carragher, B, Mancia, F. | | Deposit date: | 2020-05-17 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of arabinosyltransferase EmbB from Mycobacterium smegmatis.
Nat Commun, 11, 2020
|
|
3HNS
 
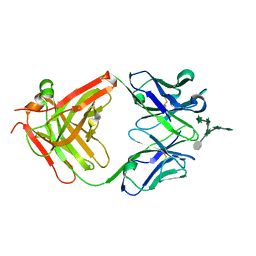 | | CS-35 Fab Complex with Oligoarabinofuranosyl Hexasaccharide | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-[beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)]alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3HNV
 
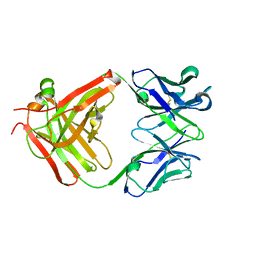 | | CS-35 Fab Complex with Oligoarabinofuranosyl Tetrasaccharide (branch part of Hexasaccharide) | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
3HNT
 
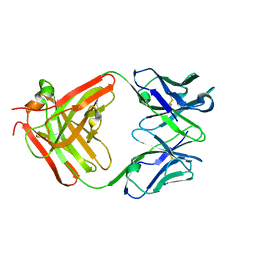 | | CS-35 Fab complex with a linear, terminal oligoarabinofuranosyl tetrasaccharide from lipoarabinomannan | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)-alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
8ZEA
 
 | |
8ZDQ
 
 | | Cryo-EM structure of Mycobacteriophage Douge complete baseplate (gp13, gp17, gp23, gp16, gp18 and gp20) | | Descriptor: | Baseplate Hub Protein (gp18), Baseplate Upper Protein (gp23), Central Fiber Protein (gp20), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDP
 
 | |
8ZDJ
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-packed connector (gp5, gp9, gp10, gp12 and gp13) | | Descriptor: | Adaptor Protein (gp9), Portal Protein (gp5), Stopper Protein (gp10), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDI
 
 | |
8ZDH
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-packed capsid (gp8 and gp113) | | Descriptor: | Capsid Cement Protein (gp113), Major Capsid Protein (gp8) | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDN
 
 | |
8ZDO
 
 | | Cryo-EM structure of Mycobacteriophage Douge baseplate (gp13, gp17, gp23, gp16, gp18 and gp20) | | Descriptor: | Baseplate hub protein (gp18), Baseplate upper protein (gp23), Central fiber protein (gp20), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDL
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-free connector (gp5, gp9, gp10, gp12 and gp13) | | Descriptor: | Adaptor Protein (gp9), Protal Protein (gp5), Stopper Protein (gp10), ... | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
8ZDM
 
 | |
8ZDK
 
 | | Cryo-EM structure of Mycobacteriophage Douge genome-packed vertex (gp8 and gp113) | | Descriptor: | Capsid Cement Protein (gp113), Major Capsid Protein (gp8) | | Authors: | Maharana, J, Wang, C.H, Tsai, L.A, Lowary, T.L, Ho, M.C. | | Deposit date: | 2024-05-02 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mycobacteriophage cryo-EM studies reveal the siphophage architecture at the amino acid level and its host interaction for viral genome ejection
To Be Published
|
|
2W5N
 
 | | Native structure of the GH93 alpha-L-arabinofuranosidase of Fusarium graminearum | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-L-ARABINOFURANOSIDASE, GLYCEROL, ... | | Authors: | Carapito, R, Imberty, A, Jeltsch, J.M, Byrns, S.C, Tam, P.H, Lowary, T.L, Varrot, A, Phalip, V. | | Deposit date: | 2008-12-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis of Arabinobio-Hydrolase Activity in Phytopathogenic Fungi. Crystal Structure and Catalytic Mechanism of Fusarium Graminearum Gh93 Exo-Alpha-L-Arabinanase.
J.Biol.Chem., 284, 2009
|
|
