5YBB
 
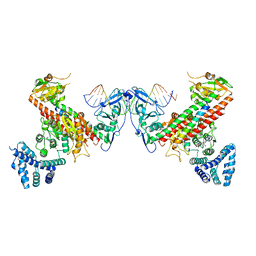 | | Structural basis underlying complex assembly andconformational transition of the type I R-M system | | Descriptor: | DNA, Restriction endonuclease S subunits, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, Y.P, Tang, Q, Zhang, J.Z, Tian, L.F, Gao, P, Yan, X.X. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis underlying complex assembly and conformational transition of the type I R-M system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7D3V
 
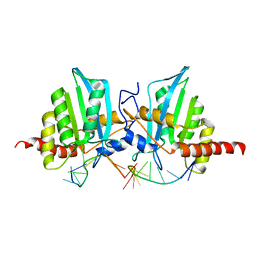 | |
4O6P
 
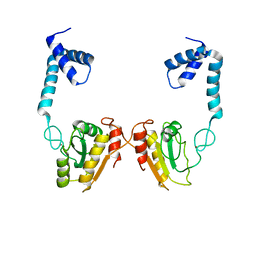 | | Structural and functional studies the characterization of C58G/C70G mutant in Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
4O6O
 
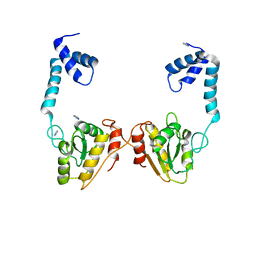 | | Structural and functional studies the characterization of Cys4 Zinc-finger motif in the recombination mediator protein RecR | | Descriptor: | IMIDAZOLE, Recombination protein RecR, ZINC ION | | Authors: | Tang, Q, Liu, Y.P, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-12-23 | | Release date: | 2014-12-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and functional characterization of Cys4 zinc finger motif in the recombination mediator protein RecR.
DNA Repair (Amst.), 24, 2014
|
|
5YDE
 
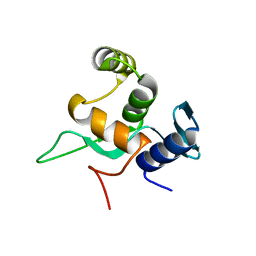 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.023 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
5YDF
 
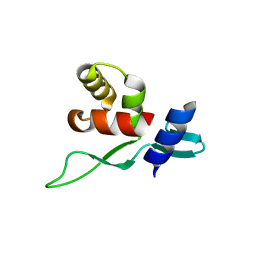 | | Crystal structure of a disease-related gene, hCDC73(1-100) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
4LU9
 
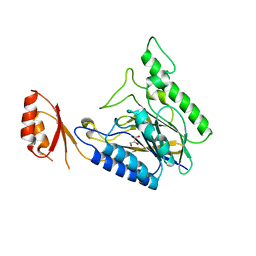 | | Crystal structure of E.coli SbcD at 2.5 angstrom resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M0V
 
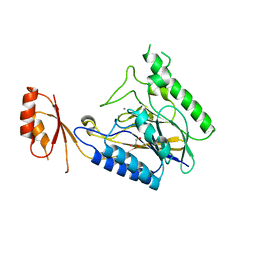 | | Crystal structure of E.coli SbcD with Mn2+ | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL, MANGANESE (II) ION | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LTY
 
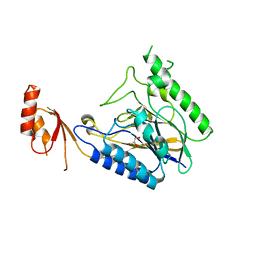 | | Crystal Structure of E.coli SbcD at 1.8 A Resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3VE5
 
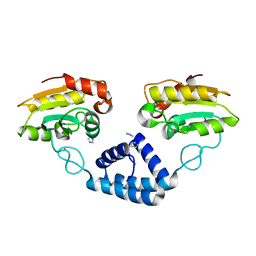 | |
3VDU
 
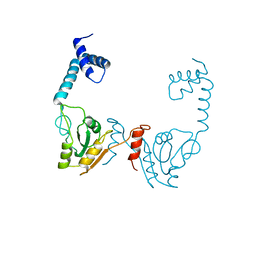 | |
3VDP
 
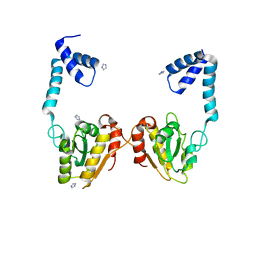 | |
4G8B
 
 | |
4G9E
 
 | |
4G5X
 
 | | Crystal structures of N-acyl homoserine lactonase AidH | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Liang, D.C, Yan, X.X, Gao, A. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High-resolution structures of AidH complexes provide insights into a novel catalytic mechanism for N-acyl homoserine lactonase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G8C
 
 | |
4G8D
 
 | |
4G9G
 
 | |
6IE2
 
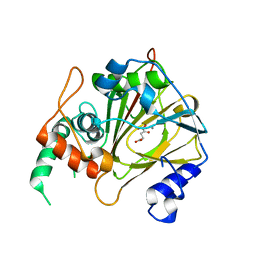 | | Crystal structure of methyladenine demethylase | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
6IE3
 
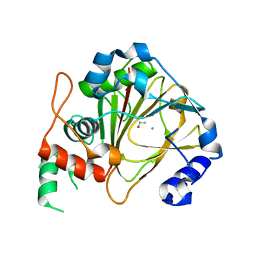 | | Crystal structure of methyladenine demethylase | | Descriptor: | ETHANOL, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
6JDE
 
 | | crystal structure of a DNA repair protein | | Descriptor: | Putative DNA repair helicase RadD, ZINC ION | | Authors: | Yan, X.X, Tang, Q. | | Deposit date: | 2019-02-01 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel ATPase RadD from Escherichia coli.
Proteins, 87, 2019
|
|
5Z68
 
 | | Structure of the recombination mediator protein RecF-ATP in RecFOR pathway | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA replication and repair protein RecF, IMIDAZOLE, ... | | Authors: | Tang, Q, Liu, Y.-P, Yan, X.-X. | | Deposit date: | 2018-01-22 | | Release date: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP-dependent conformational change in ABC-ATPase RecF serves as a switch in DNA repair.
Sci Rep, 8, 2018
|
|
5Z69
 
 | |
5Z67
 
 | |
