6PM9
 
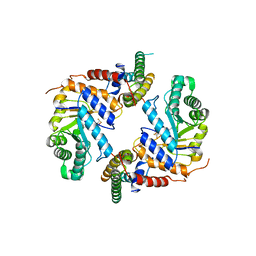 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
8J7Y
 
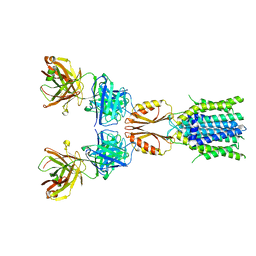 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7U
 
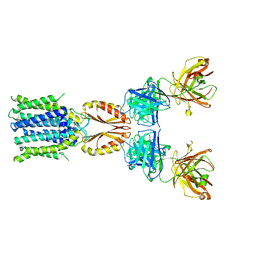 | | Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7V
 
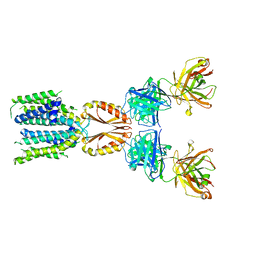 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7T
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J80
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7W
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7X
 
 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
5TRL
 
 | |
5TRM
 
 | |
7RRB
 
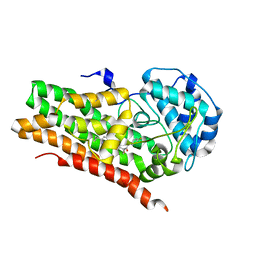 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | Descriptor: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRC
 
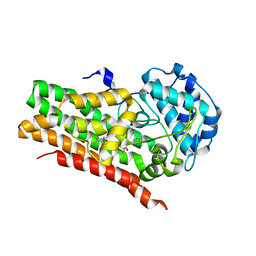 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | Authors: | Lesburg, C.A. | | Deposit date: | 2021-08-09 | | Release date: | 2022-03-16 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
6WLG
 
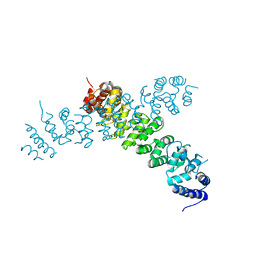 | | Ints3 C-terminal Domain | | Descriptor: | Integrator complex subunit 3 | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2020-04-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.111 Å) | | Cite: | Structural basis for multifunctional roles of human Ints3 C-terminal domain.
J.Biol.Chem., 296, 2020
|
|
6WPE
 
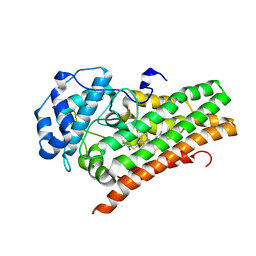 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5CMZ
 
 | | Artificial HIV fusion inhibitor AP3 fused to the C-terminus of gp41 NHR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, Artificial HIV entry inhibitor AP3, ... | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CN0
 
 | | Artificial HIV fusion inhibitor AP2 fused to the C-terminus of gp41 NHR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Envelope glycoprotein,AP2, MAGNESIUM ION | | Authors: | Zhu, Y, Ye, S, Zhang, R. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Improved Pharmacological and Structural Properties of HIV Fusion Inhibitor AP3 over Enfuvirtide: Highlighting Advantages of Artificial Peptide Strategy.
Sci Rep, 5, 2015
|
|
5CMU
 
 | |
4O46
 
 | | 14-3-3-gamma in complex with influenza NS1 C-terminal tail phosphorylated at S228 | | Descriptor: | 14-3-3 protein gamma, Nonstructural protein 1, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Liu, Y, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
8WLR
 
 | |
8WLO
 
 | |
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8KC2
 
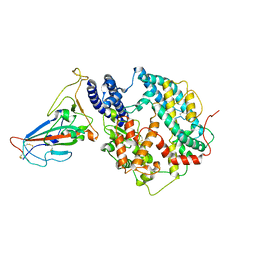 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8KA8
 
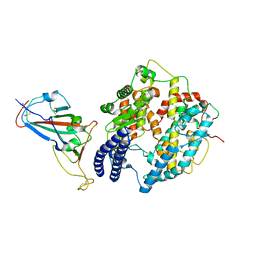 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
4NW2
 
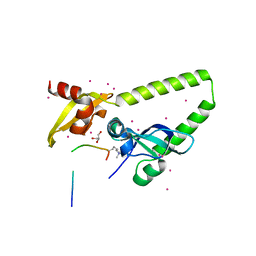 | | Tandem chromodomains of human CHD1 in complex with Influenza virus NS1 C-terminal tail trimethylated at K229 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, GLYCEROL, Nonstructural protein 1, ... | | Authors: | Qin, S, Tempel, W, Xu, C, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-05 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
