3UGQ
 
 | | Crystal structure of the apo form of the yeast mitochondrial threonyl-tRNA synthetase determined at 2.1 Angstrom resolution | | Descriptor: | POTASSIUM ION, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UH0
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with threonyl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Threonyl-tRNA synthetase, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UGT
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase - orthorhombic crystal form | | Descriptor: | Threonyl-tRNA synthetase, mitochondrial, ZINC ION | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Cho, C, Soll, D, Simonovic, M. | | Deposit date: | 2011-11-02 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Yeast mitochondrial threonyl-tRNA synthetase recognizes tRNA isoacceptors by distinct mechanisms and promotes CUN codon reassignment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EO4
 
 | | Crystal structure of the yeast mitochondrial threonyl-tRNA synthetase (MST1) in complex with seryl sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, Threonine--tRNA ligase, mitochondrial, ... | | Authors: | Peterson, K.M, Ling, J, Simonovic, I, Soll, D, Simonovic, M. | | Deposit date: | 2012-04-13 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase.
J.Biol.Chem., 287, 2012
|
|
6SJM
 
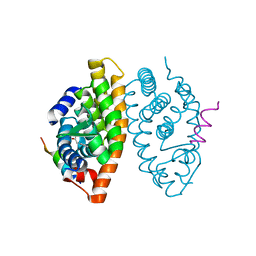 | | Crystal structure of the Retinoic Acid Receptor alpha in complex with compound 24 (JP175) | | Descriptor: | 2-[4-[3,5-bis(trifluoromethyl)phenyl]phenyl]ethanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A Novel Biphenyl-based Chemotype of Retinoid X Receptor Ligands Enables Subtype and Heterodimer Preferences.
Acs Med.Chem.Lett., 10, 2019
|
|
1YBJ
 
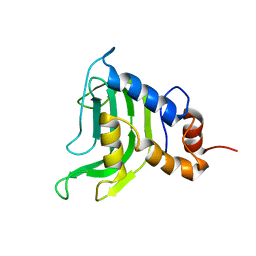 | | Structural and Dynamics studies of both apo and holo forms of the hemophore HasA | | Descriptor: | Hemophore HasA | | Authors: | Wolff, N, Izadi-Pruneyre, N, Couprie, J, Habeck, M, Linge, J, Rieping, W, Wandersman, C, Nilges, M, Delepierre, M, Lecroisey, A. | | Deposit date: | 2004-12-21 | | Release date: | 2005-12-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparative analysis of structural and dynamic properties of the loaded and unloaded hemophore HasA: functional implications.
J.Mol.Biol., 376, 2008
|
|
8A00
 
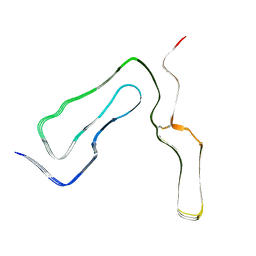 | | Infectious mouse-adapted ME7 scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2022-05-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural basis for prion strain diversity.
Nat.Chem.Biol., 19, 2023
|
|
4YE8
 
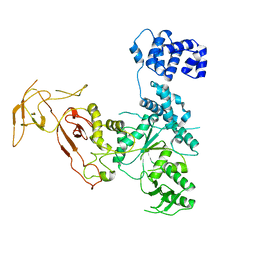 | |
4YE6
 
 | |
4YE9
 
 | |
5EUL
 
 | | Structure of the SecA-SecY complex with a translocating polypeptide substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AYC08, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Li, L, Park, E, Ling, J, Ingram, J, Ploegh, H, Rapoport, T.A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of a substrate-engaged SecY protein-translocation channel.
Nature, 531, 2016
|
|
6X7Q
 
 | | Chloramphenicol acetyltransferase type III in complex with chloramphenicol and acetyl-oxa(dethia)-CoA | | Descriptor: | CHLORAMPHENICOL, Chloramphenicol acetyltransferase 3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
6X7R
 
 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) in complex with oxa(dethia)-coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2020-05-30 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-keto-acylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(de-thia)CoA
Acta Crystallogr.,Sect.F, 2023
|
|
8D1U
 
 | | E. coli beta-ketoacyl-[acyl carrier protein] synthase III (FabH) with an acetylated cysteine and in complex with oxa(dethia)-Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, CHLORIDE ION, oxa(dethia)-CoA | | Authors: | Benjamin, A.B, Stunkard, L.M, Ling, J, Nice, J.N, Lohman, J.R. | | Deposit date: | 2022-05-27 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.302 Å) | | Cite: | Structures of chloramphenicol acetyltransferase III and Escherichia coli beta-ketoacylsynthase III co-crystallized with partially hydrolysed acetyl-oxa(dethia)CoA.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7QIG
 
 | | Infectious mouse-adapted RML scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Zhang, W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | 2.7 angstrom cryo-EM structure of ex vivo RML prion fibrils.
Nat Commun, 13, 2022
|
|
5M1X
 
 | | Crystal structure of S. cerevisiae Rfa1 N-OB domain mutant (K45E) | | Descriptor: | Replication factor A protein 1 | | Authors: | Seeber, A, Hegnauer, A.M, Hustedt, N, Deshpande, I, Poli, J, Eglinger, J, Pasero, P, Gut, H, Shinohara, M, Hopfner, K.P, Shimada, K, Gasser, S.M. | | Deposit date: | 2016-10-11 | | Release date: | 2016-12-07 | | Last modified: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPA Mediates Recruitment of MRX to Forks and Double-Strand Breaks to Hold Sister Chromatids Together.
Mol. Cell, 64, 2016
|
|
8ATY
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with JP85 (compound 1). | | Descriptor: | 2-[4-chloranyl-6-(5,6,7,8-tetrahydronaphthalen-1-ylamino)pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8PP0
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with JP147 | | Descriptor: | 3-[4-[2,3-dihydro-1H-inden-4-yl(methyl)amino]-6-(trifluoromethyl)pyrimidin-2-yl]oxypropanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Pollinger, J, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of a Highly Potent Partial RXR Agonist with Superior Physicochemical Properties.
J.Med.Chem., 67, 2024
|
|
2W9D
 
 | | Structure of Fab fragment of the ICSM 18 - anti-Prp therapeutic antibody at 1.57 A resolution. | | Descriptor: | CALCIUM ION, ICSM 18-ANTI-PRP THERAPEUTIC FAB HEAVY CHAIN, ICSM 18-ANTI-PRP THERAPEUTIC FAB LIGHT CHAIN | | Authors: | Antonyuk, S.V, Trevitt, C.R, Strange, R.W, Jackson, G.S, Sangar, D, Batchelor, M, Jones, S, Georgiou, T, Cooper, S, Fraser, C, Khalili-Shirazi, A, Clarke, A.R, Hasnain, S.S, Collinge, J. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of Human Prion Protein Bound to a Therapeutic Antibody.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W9E
 
 | | Structure of ICSM 18 (anti-Prp therapeutic antibody) Fab fragment complexed with human Prp fragment 119-231 | | Descriptor: | ICSM 18-ANTI-PRP THERAPEUTIC FAB HEAVY CHAIN, ICSM 18-ANTI-PRP THERAPEUTIC FAB LIGHT CHAIN, MAJOR PRION PROTEIN, ... | | Authors: | Antonyuk, S.V, Trevitt, C.R, Strange, R.W, Jackson, G.S, Sangar, D, Batchelor, M, Jones, S, Georgiou, T, Cooper, S, Fraser, C, Khalili-Shirazi, A, Clarke, A.R, Hasnain, S.S, Collinge, J. | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Prion Protein Bound to a Therapeutic Antibody.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2M7P
 
 | | RXFP1 utilises hydrophobic moieties on a signalling surface of the LDLa module to mediate receptor activation | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Relaxin receptor 1 | | Authors: | Kong, R.CK, Petrie, E.J, Mohanty, B, Ling, J, Lee, J.C.Y, Gooley, P.R, Bathgate, R.A.D. | | Deposit date: | 2013-04-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation.
J.Biol.Chem., 288, 2013
|
|
1WCO
 
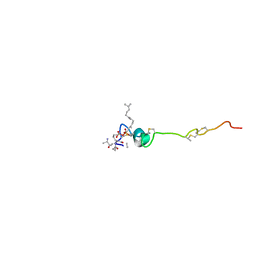 | | The solution structure of the nisin-lipid II complex | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, ALA-FGA-LYS-DAL-DAL PEPTIDE, ... | | Authors: | Hsu, S.-T.D, Breukink, E, Tischenko, E, Lutters, M.A.G, de Kruijff, B, Kaptein, R, Bonvin, A.M.J.J, van Nuland, N.A.J. | | Deposit date: | 2004-11-19 | | Release date: | 2005-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics.
Nat. Struct. Mol. Biol., 11, 2004
|
|
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
5MG0
 
 | | Structure of PAS-GAF fragment of Deinococcus phytochrome by serial femtosecond crystallography | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, ... | | Authors: | Burgie, E.S, Fuller, F.D, Gul, S, Miller, M.D, Young, I.D, Brewster, A.S, Clinger, J, Aller, P, Braeuer, P, Hutchison, C, Alonso-Mori, R, Kern, J, Yachandra, V.K, Yano, J, Sauter, N.K, Phillips Jr, G.N, Vierstra, R.D, Orville, A.M. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers.
Nat. Methods, 14, 2017
|
|
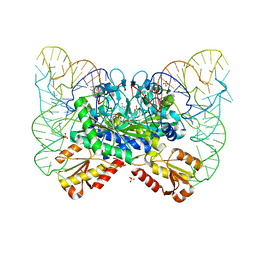
4YYE
 
 | |
