1R5D
 
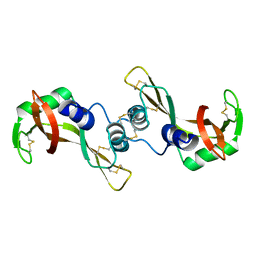 | | X-ray structure of bovine seminal ribonuclease swapping dimer from a new crystal form | | Descriptor: | Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
3BCO
 
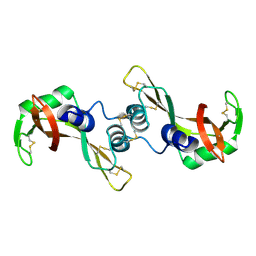 | | Crystal Structure of The Swapped FOrm of P19A/L28Q/N67D BS-RNase | | Descriptor: | Seminal ribonuclease | | Authors: | Merlino, A, Ercole, C, Picone, D, Pizzo, E, Mazzarella, L, Sica, F. | | Deposit date: | 2007-11-13 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The buried diversity of bovine seminal ribonuclease: shape and cytotoxicity of the swapped non-covalent form of the enzyme
J.Mol.Biol., 376, 2008
|
|
2PZH
 
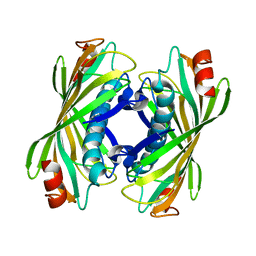 | | YbgC thioesterase (Hp0496) from Helicobacter pylori | | Descriptor: | Hypothetical protein HP_0496 | | Authors: | Angelini, A, Cendron, L, Goncalves, S, Zanotti, G, Terradot, L. | | Deposit date: | 2007-05-18 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori.
Proteins, 72, 2008
|
|
7AZT
 
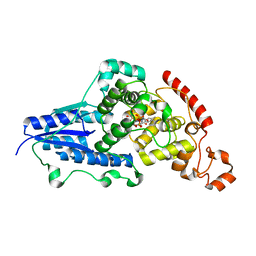 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AYV
 
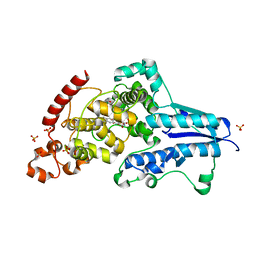 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at cryogenic temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p, ... | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3LJ9
 
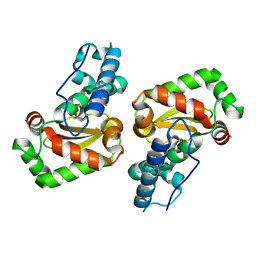 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis in complex with sodium azide | | Descriptor: | AZIDE ION, FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LIO
 
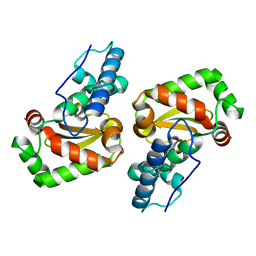 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis (crystal form I) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-25 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
3LJF
 
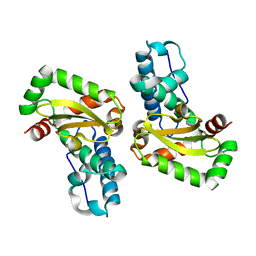 | | The X-ray structure of iron superoxide dismutase from Pseudoalteromonas haloplanktis (crystal form II) | | Descriptor: | FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, iron superoxide dismutase | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
7B6E
 
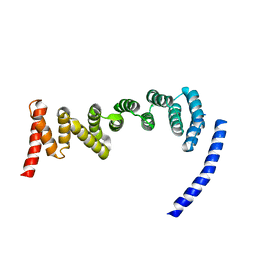 | | Drosophila melanogaster TRAPP C8 subunits region 355 to 661 | | Descriptor: | FI18195p1 | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6R
 
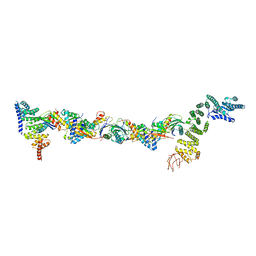 | | Drosophila melanogaster TRAPPIII partial complex: core plus C8 and C11 attached region | | Descriptor: | FI18195p1, GEO08327p1, Probable trafficking protein particle complex subunit 2, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-08 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6D
 
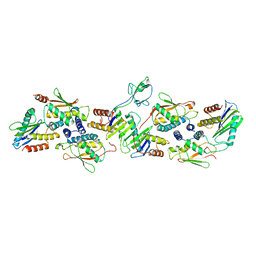 | | Drosophila melanogaster TRAPPCore (C1, C2, C2L, C3a/b, C4, C5, C6 subunits) | | Descriptor: | GEO08327p1, Probable trafficking protein particle complex subunit 2, TRAPPC2L, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6H
 
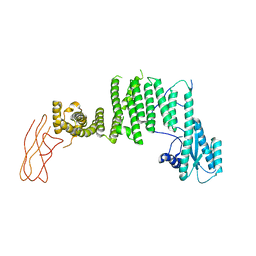 | | Drosophila melanogaster TRAPP C11 subunits region 1 to 718 | | Descriptor: | Trafficking protein particle complex subunit 11 | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J, Degliesposti, G. | | Deposit date: | 2020-12-07 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B70
 
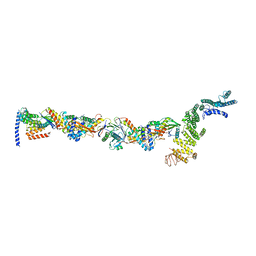 | | TRAPPCore plus C8 (355-596) and C11 (1-718) from MiniTRAPPIII | | Descriptor: | FI18195p1, GEO08327p1, Probable trafficking protein particle complex subunit 2, ... | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of metazoan TRAPPIII, the multi-subunit complex that activates the GTPase Rab1.
Embo J., 40, 2021
|
|
7B6X
 
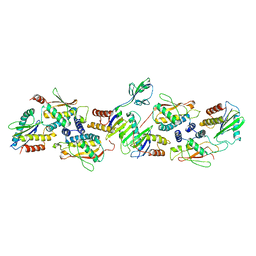 | |
7B6Y
 
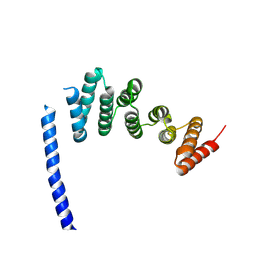 | |
7B6Z
 
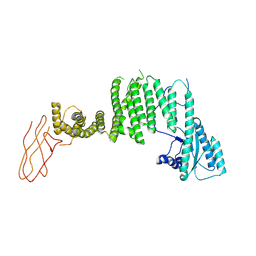 | | TRAPPC11 subunit (1-716) | | Descriptor: | Trafficking protein particle complex subunit 11 | | Authors: | Galindo, A, Munro, S, Planelles-Herrero, V.J. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural insights into the metazoan TRAPP complexes that activate Rab1 and
Rab11.
To Be Published
|
|
1R5C
 
 | | X-ray structure of the complex of Bovine seminal ribonuclease swapping dimer with d(CpA) | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, Ribonuclease, seminal | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Population shift vs induced fit: The case of bovine seminal ribonuclease swapping dimer
Biopolymers, 73, 2004
|
|
6CHA
 
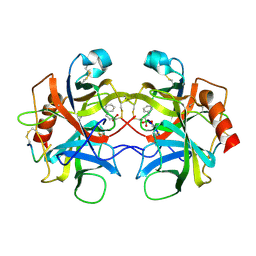 | |
6FF5
 
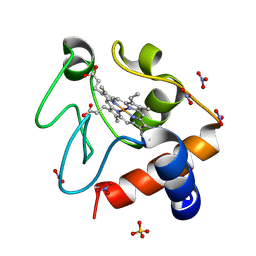 | |
3GKV
 
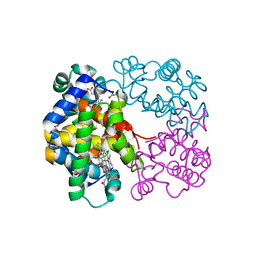 | | X-ray structure of an intermediate along the oxidation pathway of Trematomus bernacchii hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Merlino, A, Vitagliano, L, Sica, F, Vergara, A, Mazzarella, L. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Combined crystallographic and spectroscopic analysis of Trematomus bernacchii hemoglobin highlights analogies and differences in the peculiar oxidation pathway of Antarctic fish hemoglobins
Biopolymers, 91, 2009
|
|
7QUT
 
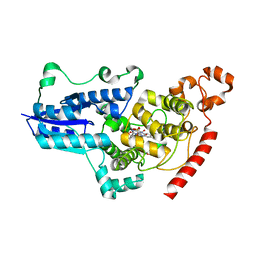 | | serial synchrotron crystallographic structure of Drosophila Melanogaster (6-4) photolyase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, RE11660p | | Authors: | Cellini, A, Weixiao, Y.W, Kumar, M.S, Westenhoff, S. | | Deposit date: | 2022-01-18 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the radical pair state in photolyases and cryptochromes.
Chem.Commun.(Camb.), 58, 2022
|
|
5EE5
 
 | | Structure of human ARL1 in complex with the DCB domain of BIG1 | | Descriptor: | ACETATE ION, ADP-ribosylation factor-like protein 1, Brefeldin A-inhibited guanine nucleotide-exchange protein 1, ... | | Authors: | Galindo, A, Soler, N, Munro, S. | | Deposit date: | 2015-10-22 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structural Insights into Arl1-Mediated Targeting of the Arf-GEF BIG1 to the trans-Golgi.
Cell Rep, 16, 2016
|
|
1YV4
 
 | | X-ray structure of M23L onconase at 100K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV7
 
 | | X-ray structure of (C87S,des103-104) onconase | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV6
 
 | | X-ray structure of M23L onconase at 298K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
