9DTQ
 
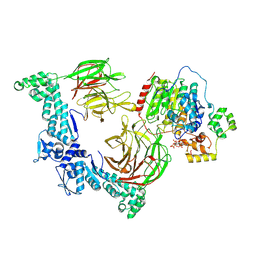 | |
8VPQ
 
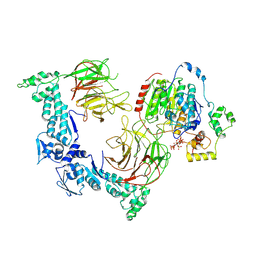 | |
8VRT
 
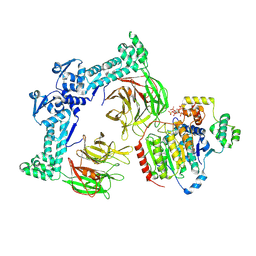 | |
8VOJ
 
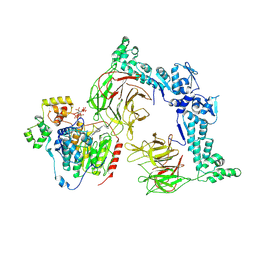 | | The Cryo-EM structure of LSD1-CoREST-HDAC1 in complex with KBTBD4 enhanced by UM171 and IP6 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-01-15 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Asymmetric Engagement of Dimeric CRL3KBTBD4 by the Molecular Glue UM171 Licenses Degradation of HDAC1/2 Complexes
To Be Published
|
|
9DTG
 
 | | The cryo-EM structure of apo KBTBD4 | | Descriptor: | Isoform 2 of Kelch repeat and BTB domain-containing protein 4 | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-09-30 | | Release date: | 2024-11-27 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Nature, 639, 2025
|
|
5V8C
 
 | | LytR-Csp2A-Psr enzyme from Actinomyces oris | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Amer, B.R, Sawaya, M.R, Liauw, B, Fu, J.Y, Ton-That, H, Clubb, R.T. | | Deposit date: | 2017-03-21 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein.
MBio, 10, 2019
|
|
