4M2J
 
 | | Crystal structure of PLP-dependent cyclase OrfR in complex with Au | | Descriptor: | Aminotransferase, GOLD ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MFK
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA | | Descriptor: | Putative uncharacterized protein tcp24, SULFATE ION, decanoyl-CoA | | Authors: | Lyu, S.Y, Liu, Y.C, Chang, C.Y, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4M2C
 
 | | Crystal structure of non-heme iron oxygenase OrfP in complex with Fe and D-Arg | | Descriptor: | D-ARGININE, FE (III) ION, L-arginine beta-hydroxylase | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-08-05 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4M74
 
 | | Mutant structure of methyltransferase from Streptomyces hygroscopicus | | Descriptor: | (2R)-2-hydroxy-3-phenylpropanoic acid, (2S,3R)-2,3-dihydroxy-3-phenylpropanoic acid, CALCIUM ION, ... | | Authors: | Liu, Y.C, Zou, X.W, Chan, H.C, Huang, C.J, Li, T.L. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of a nonhaem-iron SAM-dependent C-methyltransferase and its engineering to a hydratase and an O-methyltransferase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7XQA
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXR
 
 | | Orf1 R342A-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XX0
 
 | | C281S glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XYE
 
 | | The apo structure of Orf1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.482 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXC
 
 | | Orf1-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7Y0X
 
 | | Orf1-glycine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXD
 
 | | Orf1-sarcosine complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-formimidoyl fortimicin A synthase, SARCOSINE | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-29 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XYL
 
 | | E426Q-glycine-glycylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-06-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXM
 
 | | Orf1-glycine-4-aminobutylthricin complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7XXP
 
 | | F316A-glycine-streptothricin F complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4NE0
 
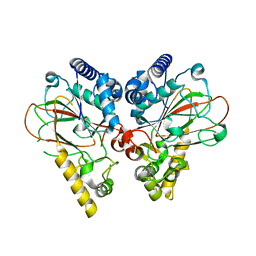 | | Crystal structure of non-heme iron oxygenase OrfP D157A mutant in complex with (3S)-hydroxy-L-Arg | | Descriptor: | (2S,3S)-3-HYDROXYARGININE, FE (III) ION, L-arginine beta-hydroxylase, ... | | Authors: | Chang, C.Y, Liu, Y.C, Lyu, S.Y, Wu, C.C, Li, T.L. | | Deposit date: | 2013-10-28 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Biosynthesis of streptolidine involved two unexpected intermediates produced by a dihydroxylase and a cyclase through unusual mechanisms
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8IZL
 
 | |
8IZM
 
 | |
4Q38
 
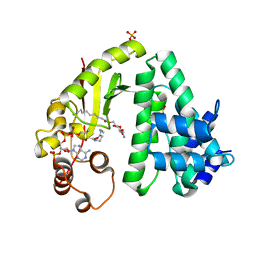 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and teicoplanin | | Descriptor: | 2-amino-6-O-decanoyl-2-deoxy-beta-D-glucopyranose, COENZYME A, Putative uncharacterized protein tcp24, ... | | Authors: | Li, T.-L, Lyu, S.-Y, Liu, Y.-C, Chang, C.-Y, Huang, C.-J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4Q36
 
 | | The crystal structure of acyltransferase in complex with octanoyl-CoA and teicoplanin | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, OCTANOYL-COENZYME A, Putative uncharacterized protein tcp24, ... | | Authors: | Li, T.-L, Lyu, S.-Y, Liu, Y.-C, Chang, C.-Y, Huang, C.-J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
7F0A
 
 | |
7F0C
 
 | | Crystal structure of capreomycin phosphotransferase in complex with CMN IIA | | Descriptor: | Capreomycin phosphotransferase, DPP-SER-DPP-UAL-MYN-KBE | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
7F0B
 
 | | Crystal structure of capreomycin phosphotransferase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capreomycin phosphotransferase | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
7F0F
 
 | | Crystal structure of capreomycin phosphotransferase in complex with CMN IIB | | Descriptor: | Capreomycin phosphotransferase, DPP-ALA-DPP-UAL-MYN-KBE | | Authors: | Chang, C.Y, Pan, Y.C, Wang, Y.L, Toh, S.I. | | Deposit date: | 2021-06-03 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual-Mechanism Confers Self-Resistance to the Antituberculosis Antibiotic Capreomycin.
Acs Chem.Biol., 17, 2022
|
|
5X8F
 
 | |
5X8G
 
 | |
