6NYF
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 1 (OA-1) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYM
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2d (OA-2d) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYJ
 
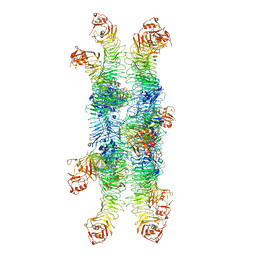 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2b (OA-2b) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYN
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2e (OA-2e) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NYL
 
 | | Helicobacter pylori Vacuolating Cytotoxin A Oligomeric Assembly 2c (OA-2c) | | Descriptor: | Vacuolating cytotoxin autotransporter | | Authors: | Zhang, K, Zhang, H, Li, S, Au, S, Chiu, W. | | Deposit date: | 2019-02-11 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures ofHelicobacter pylorivacuolating cytotoxin A oligomeric assemblies at near-atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6VQV
 
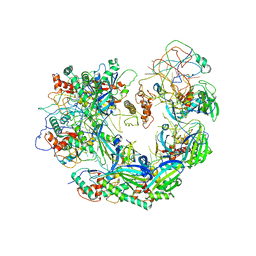 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1OOJ
 
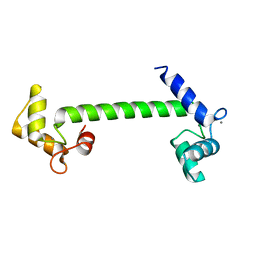 | | Structural genomics of Caenorhabditis elegans : Calmodulin | | Descriptor: | CALCIUM ION, Calmodulin CMD-1 | | Authors: | Symersky, J, Lin, G, Li, S, Qiu, S, Luan, C.-H, Luo, D, Tsao, J, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural genomics of caenorhabditis elegans: crystal structure of calmodulin.
Proteins, 53, 2003
|
|
6VQX
 
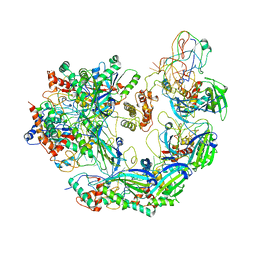 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8GTB
 
 | | Cryo-EM structure of the marine siphophage vB_DshS-R4C tail tube protein | | Descriptor: | Major tail protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTD
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C portal-adaptor complex | | Descriptor: | Head-to-tail joining protein, Portal protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTC
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C baseplate-tail complex | | Descriptor: | Distal tail protein, Hub protein, Major tail protein, ... | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
8GTF
 
 | | Cryo-EM model of the marine siphophage vB_DshS-R4C stopper-terminator complex | | Descriptor: | Head-to-tail joining protein, Major tail protein, Terminator protein | | Authors: | Huang, Y, Sun, H, Wei, S, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
1F71
 
 | |
1F70
 
 | |
1Q34
 
 | | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans | | Descriptor: | Ubiquitin-conjugating enzyme E2-21.5 kDa | | Authors: | Schormann, N, Lin, G, Li, S, Symersky, J, Qiu, S, Finley, J, Luo, D, Stanton, A, Carson, M, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-07-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two UBC (E2) enzymes of the ubiquitin-conjugating system in Caenorhabditis elegans
To be Published
|
|
1FNK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNJ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1QWK
 
 | | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa protein (aldose reductase family member) | | Descriptor: | aldo-keto reductase family 1 member C1 | | Authors: | Chen, L, Zhou, X.E, Meehan, E.J, Symersky, J, Lu, S, Li, S, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-09-02 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of Caenorhabditis Elegans: Hypothetical 35.2 kDa
protein (aldose reductase family member)
To be published
|
|
1H53
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, CITRIC ACID, PHOSPHATE ION | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
1H52
 
 | | Binding of Phosphate and Pyrophosphate ions at the active site of human Angiogenin as revealed by X-ray Crystallography | | Descriptor: | ANGIOGENIN, PYROPHOSPHATE 2- | | Authors: | Leonidas, D.D, Chavali, G.B, Jardine, A.M, Li, S, Shapiro, R, Acharya, K.R. | | Deposit date: | 2001-05-18 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of Phosphate and Pyrophosphate Ions at the Active Site of Human Angiogenin as Revealed by X-Ray Crystallography
Protein Sci., 10, 2001
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
8GTA
 
 | | Cryo-EM structure of the marine siphophage vB_Dshs-R4C capsid | | Descriptor: | Major capsid protein | | Authors: | Sun, H, Huang, Y, Zheng, Q, Li, S, Zhang, R, Xia, N. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
5V6J
 
 | | Glycan binding protein Y3 from mushroom Coprinus comatus possesses anti-leukemic activity | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, TMV resistance protein Y3 | | Authors: | Li, K, Zhang, P, Gang, Y, Xia, C, Polston, J.E, Li, G, Li, S, Lin, Z, Yang, L.-J, Bruner, S.D, Ding, Y. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Cytotoxic protein from the mushroom Coprinus comatus possesses a unique mode for glycan binding and specificity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
