5W4Y
 
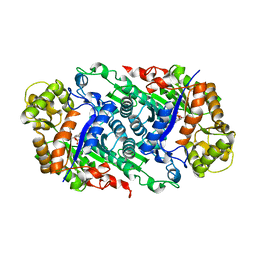 | | Crystal Structure of Riboflavin Lyase (RcaE) with cofactor FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
2AYO
 
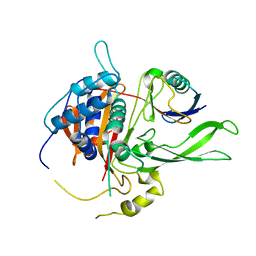 | | Structure of USP14 bound to ubquitin aldehyde | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 14 | | Authors: | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2005-09-07 | | Release date: | 2005-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
5W4Z
 
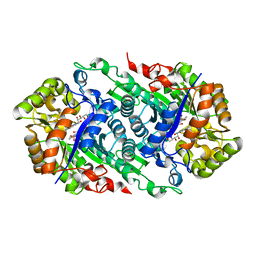 | | Crystal Structure of Riboflavin Lyase (RcaE) with modified FMN and substrate Riboflavin | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-3-O-phosphono-D-ribitol, RIBOFLAVIN, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
5W48
 
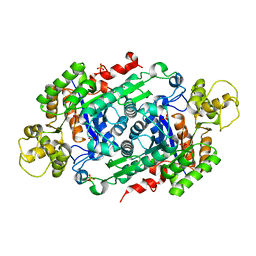 | | Crystal Structure of Riboflavin Lyase (RcaE) | | Descriptor: | Riboflavin Lyase, SULFATE ION | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
7WQY
 
 | |
7W3S
 
 | | The complex structure of Larg1-ADPr from Legionella pneumophila | | Descriptor: | Type IV secretion protein Dot, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H, Li, P. | | Deposit date: | 2021-11-26 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-18 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation.
mLife, 2022
|
|
5KEV
 
 | |
5KEW
 
 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt taurodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tomchick, D.R, Orth, K, Rivera-Cancel, G. | | Deposit date: | 2016-06-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Bile salt receptor complex activates a pathogenic type III secretion system.
Elife, 5, 2016
|
|
5B25
 
 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
3KY9
 
 | | Autoinhibited Vav1 | | Descriptor: | Proto-oncogene vav, ZINC ION | | Authors: | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.731 Å) | | Cite: | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
6X5A
 
 | | The mouse cGAS catalytic domain binding to human nucleosome that purified from HEK293T cells | | Descriptor: | Cyclic GMP-AMP synthase, DNA (natural), Histone H2A type 1, ... | | Authors: | Pengbiao, X, Pingwei, L, Baoyu, Z. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
6D38
 
 | |
7BXG
 
 | | MavC-UBE2N-Ub complex | | Descriptor: | MavC, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7BXH
 
 | | MavC-Lpg2149 complex | | Descriptor: | Lpg2149, MavC | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
4F2L
 
 | | Structure of a regulatory domain of AMPK | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, MAGNESIUM ION | | Authors: | Xin, F.J, Zhang, Y.Y, Wang, J, Wang, Z.X, Wu, J.W. | | Deposit date: | 2012-05-08 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conserved regulatory elements in AMPK
Nature, 498, 2013
|
|
4XVC
 
 | |
8JJ9
 
 | |
7LUK
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH AN AZATRICYCLIC RORGT INVERSE AGONIST | | Descriptor: | (2S)-N-[(6aS,7R,9aS)-9a-[(4-fluorophenyl)sulfonyl]-3-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-6,6a,7,8,9,9a-hexahydro-5H-cyclopenta[f]quinolin-7-yl]-2-hydroxy-2-methyl-3-(methylsulfonyl)propanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Azatricyclic Inverse Agonists of ROR gamma t That Demonstrate Efficacy in Models of Rheumatoid Arthritis and Psoriasis.
Acs Med.Chem.Lett., 12, 2021
|
|
6U25
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS- RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC INVERSE AGONIST | | Descriptor: | GLYCEROL, NUCLEAR RECEPTOR COACTIVATOR 1 CHIMERA, trans-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Rationally Designed, Conformationally Constrained Inverse Agonists of ROR gamma t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy.
J.Med.Chem., 62, 2019
|
|
5S9R
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMS-986158, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9O
 
 | | CRYSTAL STRUCTURE OF THE HUMAN BRD2 BD1 BROMODOMAIN IN COMPLEX WITH 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-(cyclopropylmethyl)-7-[(2R,6S)-2,6-dimethylmorpholine-4-carbonyl]-3-(3,5-dimethyl-1,2-oxazol-4-yl)-9H-carbazole-1-carboxamide, Bromodomain-containing protein 2 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9P
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 9-benzyl-2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9H-carbazole-4-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 9-benzyl-2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9H-carbazole-4-carboxamide, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
5S9Q
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9-[(S)-(oxan-4-yl)(phenyl)methyl]-9H-carbazole-4-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,5-dimethyl-1,2-oxazol-4-yl)-7-(2-hydroxypropan-2-yl)-9-[(S)-(oxan-4-yl)(phenyl)methyl]-9H-carbazole-4-carboxamide, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Preclinical Pharmacology of an Oral Bromodomain and Extra-Terminal (BET) Inhibitor Using Scaffold-Hopping and Structure-Guided Drug Design.
J.Med.Chem., 64, 2021
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T9U
 
 | | Crystal structure of hSTING with an agonist (SHR169224) | | Descriptor: | (3S,4S)-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-4-(prop-2-en-1-yl)-4,5-dihydroimidazo[1,5,4-de][1,4]benzoxazine-8-carboxamide, CALCIUM ION, Stimulator of interferon genes protein | | Authors: | Chowdhury, R, Miller, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | SHR1032, a novel STING agonist, stimulates anti-tumor immunity and directly induces AML apoptosis.
Sci Rep, 12, 2022
|
|
