6TIN
 
 | |
6TIB
 
 | |
6TIJ
 
 | |
6TIE
 
 | |
6TIH
 
 | |
3SIG
 
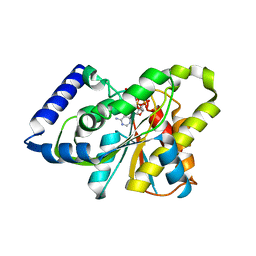 | | The X-ray crystal structure of poly(ADP-ribose) glycohydrolase (PARG) bound to ADP-ribose from Thermomonospora curvata | | Descriptor: | [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE, poly(ADP-ribose) glycohydrolase | | Authors: | Leys, D, Dunstan, M.S. | | Deposit date: | 2011-06-18 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase.
Nature, 477, 2011
|
|
7ABO
 
 | |
7ABN
 
 | | Structure of the reversible pyrrole-2-carboxylic acid decarboxylase PA0254/HudA | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, IMIDAZOLE, MANGANESE (II) ION, ... | | Authors: | Leys, D. | | Deposit date: | 2020-09-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Mechanism of Pseudomonas aeruginosa PA0254/HudA, a prFMN-Dependent Pyrrole-2-carboxylic Acid Decarboxylase Linked to Virulence.
Acs Catalysis, 11, 2021
|
|
6ZXX
 
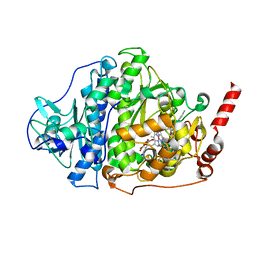 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged. | | Descriptor: | 3 bromo 4 hydroxybenzoic acid, 3,5-bis(bromanyl)-4-oxidanyl-benzoic acid, BROMIDE ION, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
6ZXU
 
 | |
6ZY1
 
 | |
6ZY0
 
 | | Catabolic reductive dehalogenase NpRdhA, N-terminally tagged, K488Q variant | | Descriptor: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | Authors: | Leys, D, Halliwell, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Catabolic Reductive Dehalogenase Substrate Complex Structures Underpin Rational Repurposing of Substrate Scope.
Microorganisms, 8, 2020
|
|
4L40
 
 | | Structure of the P450 OleT with a C20 fatty acid substrate bound | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase, icosanoic acid | | Authors: | Leys, D. | | Deposit date: | 2013-06-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
4L54
 
 | | Structure of cytochrome P450 OleT, ligand-free | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Terminal olefin-forming fatty acid decarboxylase | | Authors: | Leys, D. | | Deposit date: | 2013-06-10 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium.
J.Biol.Chem., 289, 2014
|
|
3CXY
 
 | |
3CY0
 
 | |
3CXZ
 
 | |
3CY1
 
 | |
3CXV
 
 | |
3CXX
 
 | |
3EKB
 
 | |
6TIL
 
 | |
6TIO
 
 | |
4KEW
 
 | | structure of the A82F BM3 heme domain in complex with omeprazole | | Descriptor: | 6-methoxy-2-{[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]sulfanyl}-1H-benzimidazole, Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
4KF0
 
 | | Structure of the A82F P450 BM3 heme domain | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Key Mutations Alter the Cytochrome P450 BM3 Conformational Landscape and Remove Inherent Substrate Bias.
J.Biol.Chem., 288, 2013
|
|
