6DUJ
 
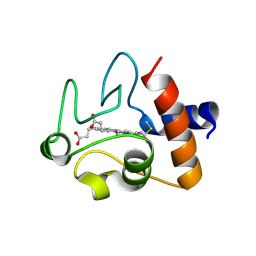 | | Crystal structure of A51V variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Lei, H, Bowler, B.E. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82202721 Å) | | Cite: | Naturally Occurring A51V Variant of Human CytochromecDestabilizes the Native State and Enhances Peroxidase Activity.
J.Phys.Chem.B, 123, 2019
|
|
6XNK
 
 | |
4PT5
 
 | |
4PSO
 
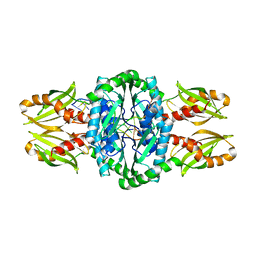 | | Crystal structure of apeThermo-DBP-RP2 bound to ssDNA dT10 | | Descriptor: | PHOSPHATE ION, polydeoxyribonucleotide, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSN
 
 | | Crystal structure of apeThermo-DBP-RP2 | | Descriptor: | GLYCEROL, IMIDAZOLE, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSM
 
 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form II) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4PSL
 
 | | Crystal structure of pfuThermo-DBP-RP1 (crystal form I) | | Descriptor: | SULFATE ION, ssDNA binding protein | | Authors: | Gahlei, H, von Moeller, H, Eppers, D, Sohmen, D, Wilson, D.N, Loll, B, Wahl, M.C. | | Deposit date: | 2014-03-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Entrapment of DNA in an intersubunit tunnel system of a single-stranded DNA-binding protein.
Nucleic Acids Res., 42, 2014
|
|
4RNA
 
 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
7MRI
 
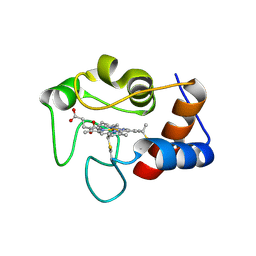 | | Crystal structure of N63T yeast iso-1-cytochrome c | | Descriptor: | Cytochrome c isoform 1, HEME C | | Authors: | Lei, H, Bowler, B.E, Evenson, G.E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Effect on intrinsic peroxidase activity of substituting coevolved residues from Omega-loop C of human cytochrome c into yeast iso-1-cytochrome c.
J.Inorg.Biochem., 232, 2022
|
|
4YIW
 
 | | DIHYDROOROTASE FROM BACILLUS ANTHRACIS WITH SUBSTRATE BOUND | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Lei, H, Santarsiero, B.D, Rice, A.J, Lee, H, Johnson, M.E. | | Deposit date: | 2015-03-02 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase).
Bioorg.Med.Chem., 24, 2016
|
|
8UIN
 
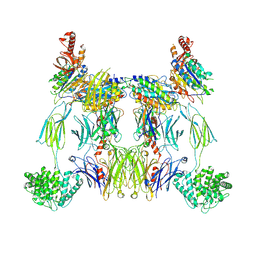 | | Structure of the C3bBb-albicin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Albicin, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Mechanism of complement inhibition by a mosquito protein revealed through cryo-EM.
Commun Biol, 7, 2024
|
|
8UH2
 
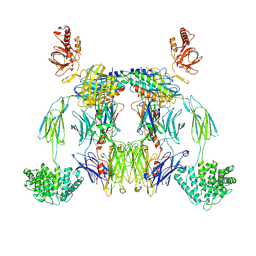 | | Complex of C3b with the inhibitor albicin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Albicin, Complement C3 beta chain, ... | | Authors: | Andersen, J.F, Lei, H, Strayer, E, Ribeiro, J.M. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of complement inhibition by a mosquito protein revealed through cryo-EM.
Commun Biol, 7, 2024
|
|
5TY3
 
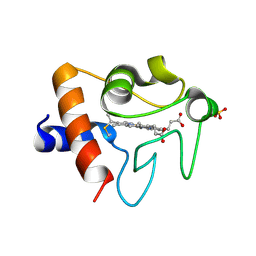 | | Crystal structure of K72A variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Mou, T.C, Nold, S.M, Lei, H, Sprang, S.R, Bowler, B.E. | | Deposit date: | 2016-11-18 | | Release date: | 2017-09-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effect of a K72A Mutation on the Structure, Stability, Dynamics, and Peroxidase Activity of Human Cytochrome c.
Biochemistry, 56, 2017
|
|
3ETA
 
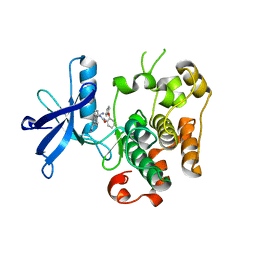 | | Kinase domain of insulin receptor complexed with a pyrrolo pyridine inhibitor | | Descriptor: | 1-(3-{5-[4-(aminomethyl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)-3-(2-phenoxyphenyl)urea, insulin receptor, kinase domain | | Authors: | Patnaik, S, Stevens, K, Gerding, R, Deanda, F, Shotwell, B, Tang, J, Hamajima, T, Nakamura, H, Leesnitzer, A, Hassell, A, Shewchuk, L, Kumar, R, Lei, H, Chamberlain, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
9C3J
 
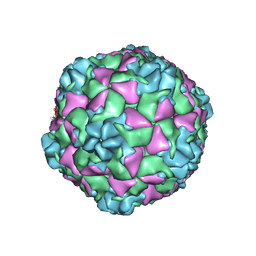 | | Cryo-EM structure of EV-D68 B3 Virus-like particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-01 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM Structure of EV-D68 Vaccine Candidate - B3 Subclade Virus-like Particle
To Be Published
|
|
9C4A
 
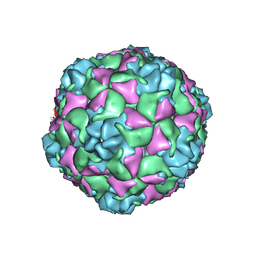 | | Cryo-EM Structure of EV-D68 Vaccine Candidate - A2 Subclade Virus-like Particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-03 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Cryo-EM Structure of EV-D68 Vaccine Candidate - A2 Subclade Virus-like Particle
To Be Published
|
|
9C8F
 
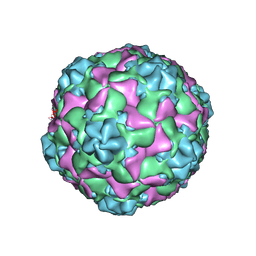 | | Cryo-EM Structure of EV-D68 B3 A-Particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Cryo-EM Structures of EV-D68 Vaccine Candidates - Virus-like Particle and Inactivated Virus Particle
To Be Published
|
|
9C8H
 
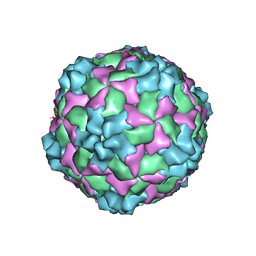 | | Cryo-EM Structure of EV-D68 A2 A-Particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM Structures of EV-D68 Vaccine Candidates - Virus-like Particle and Inactivated Virus Particle
To Be Published
|
|
9C8I
 
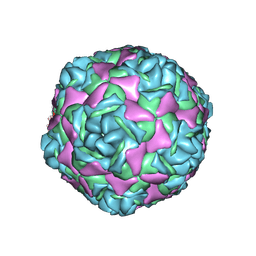 | | Cryo-EM Structure of EV-D68 B3 Inactivated Virus Particle | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM Structures of EV-D68 Vaccine Candidates - Virus-like Particle and Inactivated Virus Particle
To Be Published
|
|
9C8G
 
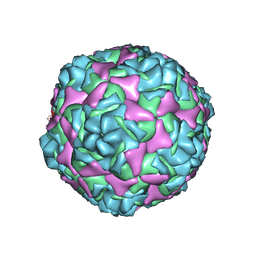 | | Cryo-EM Structure of EV-D68 A2 Inactivated Virus Particle | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Cheng, J, Krug, P.W, Lei, H, Moss, D.L, Huang, R, Lang, Z.C, Morton, A.J, Shen, C, Pierson, T.C, Zhou, T, Ruckwardt, T.J, Kwong, P.D. | | Deposit date: | 2024-06-12 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM Structures of EV-D68 Vaccine Candidates - Virus-like Particle and Inactivated Virus Particle
To Be Published
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8EOK
 
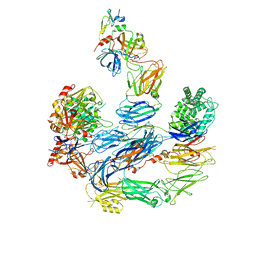 | | Structure of the C3bB proconvertase in complex with lufaxin and factor Xa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated factor Xa heavy chain, Complement C3 beta chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8ENU
 
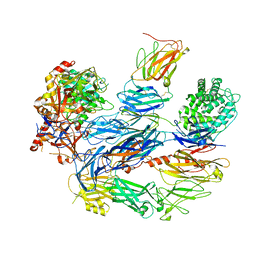 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
9COO
 
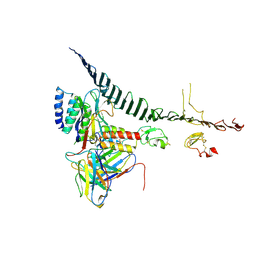 | | Nanobody 4 bound to Apolipoprotein B 100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ApoB100 nanobody 4, Apolipoprotein B 100, ... | | Authors: | Dearborn, A.D, Kumar, A, Reimund, M, Graziano, G, Lei, H, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-07-17 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
9BD1
 
 | | beta/alpha1 region of ApoB 100 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Apolipoprotein B-100 | | Authors: | Dearborn, A.D, Reimund, M, Graziano, G, Lei, H, Kumar, A, Neufeld, E.B, Remaley, A.T, Marcotrigiano, J. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of apolipoprotein B100 bound to the low-density lipoprotein receptor.
Nature, 638, 2025
|
|
