7D24
 
 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D26
 
 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | Descriptor: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D22
 
 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | Descriptor: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D25
 
 | | Hsp90 alpha N-terminal domain in complex with a 14 compund | | Descriptor: | 6-chloranyl-9-[(4-methylphenyl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D1V
 
 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | Descriptor: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | Authors: | Shin, S.C, Kim, E.E. | | Deposit date: | 2020-09-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
3HEC
 
 | | P38 in complex with Imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
3HEG
 
 | | P38 in complex with Sorafenib | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Mitogen-activated protein kinase 14 | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
3LRE
 
 | |
7FFG
 
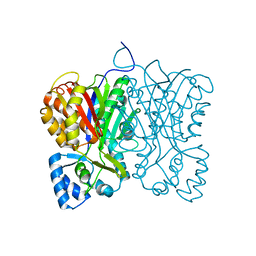 | | Diarylpentanoid-producing polyketide synthase (N199F mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFH
 
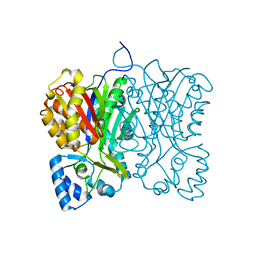 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFI
 
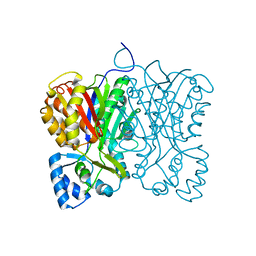 | | Diarylpentanoid-producing polyketide synthase (F340W mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFA
 
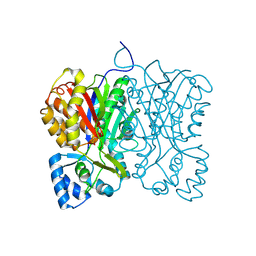 | | Diarylpentanoid-producing polyketide synthase from Aquilaria sinensis | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
7FFC
 
 | | Diarylpentanoid-producing polyketide synthase (A210E mutant) | | Descriptor: | GLYCEROL, Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Kodama, T, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
5H3C
 
 | |
5GLG
 
 | |
4LUS
 
 | | alanine racemase [Clostridium difficile 630] | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Alanine racemase, GLYCEROL | | Authors: | Asojo, O.A. | | Deposit date: | 2013-07-25 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analyses of alanine racemase from the multidrug-resistant Clostridium difficile strain 630.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2H0P
 
 | | NMR Structure of the Dengue-4 virus Envelope Protein Domain III | | Descriptor: | Envelope glycoprotein | | Authors: | Volk, D.E, Lee, Y, Li, X, Thiviyanathan, V, Barrett, A.D.T, Gorenstein, D.G. | | Deposit date: | 2006-05-15 | | Release date: | 2007-03-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the envelope protein domain III of dengue-4 virus.
Virology, 364, 2007
|
|
4LUT
 
 | | alanine racemase [Clostridium difficile 630] complex with cycloserine | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Asojo, O.A. | | Deposit date: | 2013-07-25 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and biochemical analyses of alanine racemase from the multidrug-resistant Clostridium difficile strain 630.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUY
 
 | | Crystal structure of CdALR mutant K 271 T | | Descriptor: | Alanine racemase | | Authors: | Asojo, O.A. | | Deposit date: | 2013-07-25 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analyses of alanine racemase from the multidrug-resistant Clostridium difficile strain 630.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4IXU
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 11d: {(5R)-5-amino-5-carboxy-5-[(3-endo)-8-(3,4-dichlorobenzyl)-8-azabicyclo[3.2.1]oct-3-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IXV
 
 | | Crystal structure of human Arginase-2 complexed with inhibitor 2d: {(5R)-5-amino-5-carboxy-5-[1-(4-chlorobenzyl)piperidin-4-yl]pentyl}(trihydroxy)borate(1-) | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D, Beckett, P, Van Zandt, M.C, Ji, M.K, Ryder, T, Jagdmann, R, Andreoli, M, Olczak, J, Mazur, M, Czestkowski, W, Piotrowska, W, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of quaternary alpha-amino acid-based arginase inhibitors via the Ugi reaction.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2H0F
 
 | | Crystal Structure of PucM in the presence of 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, Transthyretin-like protein pucM | | Authors: | Rhee, S. | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H0E
 
 | | Crystal Structure of PucM in the absence of substrate | | Descriptor: | GLYCEROL, Transthyretin-like protein pucM | | Authors: | Rhee, S. | | Deposit date: | 2006-05-14 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H0J
 
 | | Crystal structure of PucM in the presence of 5,6-diaminouracil | | Descriptor: | 5,6-DIAMINOPYRIMIDINE-2,4(1H,3H)-DIONE, Transthyretin-like protein pucM | | Authors: | Rhee, S. | | Deposit date: | 2006-05-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7WM5
 
 | |
